Gene
KWMTBOMO06004
Pre Gene Modal
BGIBMGA002821
Annotation
dnaJ_homolog_subfamily_C_member_16_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.79
Sequence
CDS
ATGAAATGGAACTTGAAGGGTTGGAAGGGTCGTTGGCTGTGGGCGTTGCTGGTGGTTATTCTGTGTACACTGACGGTCGCACAGAAGATCGGTGACCCATATAAAATTCTCGGTATACATCGAAAAGCAAGCTTGCCTGAGATACGAAAGGCATATCGGCAATTAGCAAAGGAATGGCACCCAGACAAGAATGAAAATCCAAATGCTGAAGACCGTTTTGTGGAGATCAAGCAAGCCTACGAGCTGCTTTCGGATACAGAACGGAGGCAGGCCTACGACTTGTATGGTATCACAAACGAAGACGACCACATGTACAAACCGAGACATGACTATAGTCAGTATGCAAGGTTCAGCAATGATCCCTTCGAAGAGTTCTTCGGCACACATTTCCGGACCCAAGATCAAGACATAACGCTCTTCCACAAACTTTCCGTCACTGCCAGACACTTCGAGAACAATATACTGGAGAAGAGCGTTCACACTCCGGCCATAGTCCTGTTCTACACGGACTGGTGCTTCGAGTGCGTGCGCGGCGCGGCGGCGTGGCGCCGGCTGGTGGAGGCCATGCAGCCGCTGGGCGTCACCATGGCGACGGTGCACGCGGGCCACGAGGCGGGCCTGGCGCGCCGCGTGGGCATCCACGGCGTGCCCTGTCTCACGCTCGTGCTCGACAAACACGTCTACGTCTACAAAGAGAGCCTGTCTTCAACGCAAAAGATATTGGAGTTCATCCGTTGGCGTTTCCCGTACAAGATGGTGTTAAGTGTGACGGATGCGAACGTGGACTCGTTCATCTCGGACTTCGAGGACAACAAAGTGAAGGCTCTGATCTTCGAGGAGCGGCACACGATGCGGCTGCGCTACCTCGTCACGGCCTTCCACTACCGCGACCGCGTGGCCTTCGGTTTCGTGGACATGAAGTCTCAAGACACGAGGAACGTGTCGGGCCGCTTCAAGGTGCAGCGCGACGTGGACACCATGGTGCTGGTGAAGGAGGACAGCGACGCGCCCGCCGCCACCGTCAGCACCGCCGAGATACCCACCGAGACACTCCACCAGCTCATCGATGCTGAACAGCTGCTGACGCTGCCCAGGCTCTCGTCGCAGGACCTCGTGTGA
Protein
MKWNLKGWKGRWLWALLVVILCTLTVAQKIGDPYKILGIHRKASLPEIRKAYRQLAKEWHPDKNENPNAEDRFVEIKQAYELLSDTERRQAYDLYGITNEDDHMYKPRHDYSQYARFSNDPFEEFFGTHFRTQDQDITLFHKLSVTARHFENNILEKSVHTPAIVLFYTDWCFECVRGAAAWRRLVEAMQPLGVTMATVHAGHEAGLARRVGIHGVPCLTLVLDKHVYVYKESLSSTQKILEFIRWRFPYKMVLSVTDANVDSFISDFEDNKVKALIFEERHTMRLRYLVTAFHYRDRVAFGFVDMKSQDTRNVSGRFKVQRDVDTMVLVKEDSDAPAATVSTAEIPTETLHQLIDAEQLLTLPRLSSQDLV
Summary
Uniprot
H6VTR5
H9IZY6
A0A2H1WUX4
A0A194Q4A0
A0A212ENP8
A0A194QR87
+ More
A0A1S4F207 Q17I06 B0WYE9 A0A1Q3F4Q7 A0A1Q3F4T5 A0A1Q3F4V5 A0A182HAZ9 A0A1L8DTT7 A0A1B0F075 U5EYA9 W5JQP5 A0A084VX54 A0A182FJI2 A0A182J0H4 A0A336KHQ9 A0A182W631 A0A1I8NFP8 A0A1I8P9H9 F7ITY4 A0A067REI9 A0A2J7RJA7 A0A2J7RJA5 B4NA00 A0A0M4EMM9 A0A0L0C205 B4KYT8 B4J264 A0A154P8B5 B4LE75 E2AQZ9 A0A2P8YH34 A0A1Y1N5Y4 A0A2A3EIQ6 D6X059 A0A182PTY3 A0A088A8H8 A0A0A9YJ78 A0A0K8T8U9 A0A310S6U1 A0A0J7KXD5 A0A195EL35 A0A195FC91 A0A0C9QAM9 E2BWJ4 A0A182W7L1 A0A0C9QVY4 A0A195CJ56 A0A151WR59 A0A1B6DHU2 A0A0R3P1Z5 A0A1J1J813 A0A026VTH3 A0A158NZJ1 A0A195BMT1 T1HM40 A0A182YQS5 A0A182MHE4 A0A182RVV9 A0A1A9YUE9 K7J2H4 A0A3B0KT24 A0A232ESD4 A0A0K8VAP4 A0A1B6K467 A0A1B6GKN3 W8BJ44 A0A182RJD8 A0A0K8UY57 A0A1A9W3K0 A0A224X855 A0A0Q5WLP3 A0A0P8XDV4 A0A3L8D825 A0A0A1WZF6 A0A0K2VET6 Q7PLG1 E0VK40 A0A087URH1 A0A1S3DS69 A0A0P4VW10 A0A1A9XZD4 A0A0P4VY93 A0A1B0BJE7 V5GFC9 Q6AWH1 A0A1S4EDK0 A0A3B0KIN6 A0A293M034 A0A0P4XQW3 A0A0N8BGB1 A0A2P2I9R6 A0A0P4XYJ1 A0A0P5NML7 A0A0P5T3I9
A0A1S4F207 Q17I06 B0WYE9 A0A1Q3F4Q7 A0A1Q3F4T5 A0A1Q3F4V5 A0A182HAZ9 A0A1L8DTT7 A0A1B0F075 U5EYA9 W5JQP5 A0A084VX54 A0A182FJI2 A0A182J0H4 A0A336KHQ9 A0A182W631 A0A1I8NFP8 A0A1I8P9H9 F7ITY4 A0A067REI9 A0A2J7RJA7 A0A2J7RJA5 B4NA00 A0A0M4EMM9 A0A0L0C205 B4KYT8 B4J264 A0A154P8B5 B4LE75 E2AQZ9 A0A2P8YH34 A0A1Y1N5Y4 A0A2A3EIQ6 D6X059 A0A182PTY3 A0A088A8H8 A0A0A9YJ78 A0A0K8T8U9 A0A310S6U1 A0A0J7KXD5 A0A195EL35 A0A195FC91 A0A0C9QAM9 E2BWJ4 A0A182W7L1 A0A0C9QVY4 A0A195CJ56 A0A151WR59 A0A1B6DHU2 A0A0R3P1Z5 A0A1J1J813 A0A026VTH3 A0A158NZJ1 A0A195BMT1 T1HM40 A0A182YQS5 A0A182MHE4 A0A182RVV9 A0A1A9YUE9 K7J2H4 A0A3B0KT24 A0A232ESD4 A0A0K8VAP4 A0A1B6K467 A0A1B6GKN3 W8BJ44 A0A182RJD8 A0A0K8UY57 A0A1A9W3K0 A0A224X855 A0A0Q5WLP3 A0A0P8XDV4 A0A3L8D825 A0A0A1WZF6 A0A0K2VET6 Q7PLG1 E0VK40 A0A087URH1 A0A1S3DS69 A0A0P4VW10 A0A1A9XZD4 A0A0P4VY93 A0A1B0BJE7 V5GFC9 Q6AWH1 A0A1S4EDK0 A0A3B0KIN6 A0A293M034 A0A0P4XQW3 A0A0N8BGB1 A0A2P2I9R6 A0A0P4XYJ1 A0A0P5NML7 A0A0P5T3I9
Pubmed
26434795
19121390
26354079
22118469
17510324
26483478
+ More
20920257 23761445 24438588 25315136 12364791 14747013 17210077 24845553 17994087 26108605 20798317 29403074 28004739 18362917 19820115 25401762 26823975 15632085 23185243 24508170 21347285 25244985 20075255 28648823 24495485 30249741 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863
20920257 23761445 24438588 25315136 12364791 14747013 17210077 24845553 17994087 26108605 20798317 29403074 28004739 18362917 19820115 25401762 26823975 15632085 23185243 24508170 21347285 25244985 20075255 28648823 24495485 30249741 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863
EMBL
JN872900
AFC01239.1
BABH01018939
BABH01018940
BABH01018941
BABH01018942
+ More
BABH01018943 ODYU01010848 SOQ56204.1 KQ459472 KPJ00174.1 AGBW02013618 OWR43122.1 KQ461175 KPJ08002.1 CH477244 EAT46270.1 DS232187 EDS37026.1 GFDL01012499 JAV22546.1 GFDL01012473 JAV22572.1 GFDL01012450 JAV22595.1 JXUM01123711 JXUM01123712 JXUM01123713 JXUM01123714 KQ566747 KXJ69854.1 GFDF01004287 JAV09797.1 AJVK01007414 AJVK01007415 GANO01000409 JAB59462.1 ADMH02000457 ETN66411.1 ATLV01017849 KE525195 KFB42548.1 UFQS01000496 UFQT01000496 SSX04404.1 SSX24768.1 AAAB01008975 EAA13497.5 KK852685 KDR18493.1 NEVH01002994 PNF40918.1 PNF40919.1 CH964232 EDW81755.2 CP012525 ALC44985.1 JRES01000994 KNC26355.1 CH933809 EDW17802.2 CH916366 EDV97015.1 KQ434844 KZC08169.1 CH940647 EDW69031.1 GL441846 EFN64145.1 PYGN01000604 PSN43484.1 GEZM01013846 JAV92300.1 KZ288254 PBC30891.1 KQ971371 EFA10042.1 GBHO01037397 GBHO01010502 GBHO01010500 GDHC01017893 JAG06207.1 JAG33102.1 JAG33104.1 JAQ00736.1 GBRD01003844 JAG61977.1 KQ767320 OAD53407.1 LBMM01002332 KMQ94968.1 KQ978747 KYN28652.1 KQ981685 KYN38028.1 GBYB01011538 GBYB01011539 JAG81305.1 JAG81306.1 GL451131 EFN79950.1 GBYB01007909 JAG77676.1 KQ977720 KYN00467.1 KQ982807 KYQ50389.1 GEDC01023083 GEDC01012103 GEDC01011267 JAS14215.1 JAS25195.1 JAS26031.1 CH475408 KRT05310.1 KRT05311.1 KRT05312.1 CVRI01000073 CRL07622.1 KK108267 EZA46801.1 ADTU01004645 KQ976433 KYM87285.1 ACPB03017303 AXCM01005472 CCAG010008949 OUUW01000014 SPP88381.1 NNAY01002440 OXU21268.1 GDHF01016689 JAI35625.1 GECU01001468 JAT06239.1 GECZ01006791 JAS62978.1 GAMC01007793 JAB98762.1 GDHF01021064 JAI31250.1 GFTR01007869 JAW08557.1 CH954177 KQS71086.1 CH902623 KPU72681.1 QOIP01000012 RLU16233.1 GBXI01010407 JAD03885.1 HACA01031035 CDW48396.1 BT021241 BT044071 AE014296 AAX33389.1 ACH92136.1 EAA46244.2 EAA46245.2 DS235239 EEB13747.1 KK121203 KFM79960.1 GDRN01104152 JAI57966.1 GDRN01104159 GDRN01104158 JAI57965.1 JXJN01015375 GALX01005787 JAB62679.1 BT015277 AAT94506.1 SPP88380.1 GFWV01008712 MAA33441.1 GDIP01239436 JAI83965.1 GDIQ01180571 JAK71154.1 IACF01005159 LAB70745.1 GDIP01237130 JAI86271.1 GDIQ01140264 JAL11462.1 GDIP01131534 JAL72180.1
BABH01018943 ODYU01010848 SOQ56204.1 KQ459472 KPJ00174.1 AGBW02013618 OWR43122.1 KQ461175 KPJ08002.1 CH477244 EAT46270.1 DS232187 EDS37026.1 GFDL01012499 JAV22546.1 GFDL01012473 JAV22572.1 GFDL01012450 JAV22595.1 JXUM01123711 JXUM01123712 JXUM01123713 JXUM01123714 KQ566747 KXJ69854.1 GFDF01004287 JAV09797.1 AJVK01007414 AJVK01007415 GANO01000409 JAB59462.1 ADMH02000457 ETN66411.1 ATLV01017849 KE525195 KFB42548.1 UFQS01000496 UFQT01000496 SSX04404.1 SSX24768.1 AAAB01008975 EAA13497.5 KK852685 KDR18493.1 NEVH01002994 PNF40918.1 PNF40919.1 CH964232 EDW81755.2 CP012525 ALC44985.1 JRES01000994 KNC26355.1 CH933809 EDW17802.2 CH916366 EDV97015.1 KQ434844 KZC08169.1 CH940647 EDW69031.1 GL441846 EFN64145.1 PYGN01000604 PSN43484.1 GEZM01013846 JAV92300.1 KZ288254 PBC30891.1 KQ971371 EFA10042.1 GBHO01037397 GBHO01010502 GBHO01010500 GDHC01017893 JAG06207.1 JAG33102.1 JAG33104.1 JAQ00736.1 GBRD01003844 JAG61977.1 KQ767320 OAD53407.1 LBMM01002332 KMQ94968.1 KQ978747 KYN28652.1 KQ981685 KYN38028.1 GBYB01011538 GBYB01011539 JAG81305.1 JAG81306.1 GL451131 EFN79950.1 GBYB01007909 JAG77676.1 KQ977720 KYN00467.1 KQ982807 KYQ50389.1 GEDC01023083 GEDC01012103 GEDC01011267 JAS14215.1 JAS25195.1 JAS26031.1 CH475408 KRT05310.1 KRT05311.1 KRT05312.1 CVRI01000073 CRL07622.1 KK108267 EZA46801.1 ADTU01004645 KQ976433 KYM87285.1 ACPB03017303 AXCM01005472 CCAG010008949 OUUW01000014 SPP88381.1 NNAY01002440 OXU21268.1 GDHF01016689 JAI35625.1 GECU01001468 JAT06239.1 GECZ01006791 JAS62978.1 GAMC01007793 JAB98762.1 GDHF01021064 JAI31250.1 GFTR01007869 JAW08557.1 CH954177 KQS71086.1 CH902623 KPU72681.1 QOIP01000012 RLU16233.1 GBXI01010407 JAD03885.1 HACA01031035 CDW48396.1 BT021241 BT044071 AE014296 AAX33389.1 ACH92136.1 EAA46244.2 EAA46245.2 DS235239 EEB13747.1 KK121203 KFM79960.1 GDRN01104152 JAI57966.1 GDRN01104159 GDRN01104158 JAI57965.1 JXJN01015375 GALX01005787 JAB62679.1 BT015277 AAT94506.1 SPP88380.1 GFWV01008712 MAA33441.1 GDIP01239436 JAI83965.1 GDIQ01180571 JAK71154.1 IACF01005159 LAB70745.1 GDIP01237130 JAI86271.1 GDIQ01140264 JAL11462.1 GDIP01131534 JAL72180.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000008820
UP000002320
+ More
UP000069940 UP000249989 UP000092462 UP000000673 UP000030765 UP000069272 UP000075880 UP000075920 UP000095301 UP000095300 UP000007062 UP000027135 UP000235965 UP000007798 UP000092553 UP000037069 UP000009192 UP000001070 UP000076502 UP000008792 UP000000311 UP000245037 UP000242457 UP000007266 UP000075885 UP000005203 UP000036403 UP000078492 UP000078541 UP000008237 UP000078542 UP000075809 UP000001819 UP000183832 UP000053097 UP000005205 UP000078540 UP000015103 UP000076408 UP000075883 UP000075900 UP000092444 UP000002358 UP000268350 UP000215335 UP000091820 UP000008711 UP000007801 UP000279307 UP000000803 UP000009046 UP000054359 UP000079169 UP000092443 UP000092460
UP000069940 UP000249989 UP000092462 UP000000673 UP000030765 UP000069272 UP000075880 UP000075920 UP000095301 UP000095300 UP000007062 UP000027135 UP000235965 UP000007798 UP000092553 UP000037069 UP000009192 UP000001070 UP000076502 UP000008792 UP000000311 UP000245037 UP000242457 UP000007266 UP000075885 UP000005203 UP000036403 UP000078492 UP000078541 UP000008237 UP000078542 UP000075809 UP000001819 UP000183832 UP000053097 UP000005205 UP000078540 UP000015103 UP000076408 UP000075883 UP000075900 UP000092444 UP000002358 UP000268350 UP000215335 UP000091820 UP000008711 UP000007801 UP000279307 UP000000803 UP000009046 UP000054359 UP000079169 UP000092443 UP000092460
Interpro
IPR036869
J_dom_sf
+ More
IPR013766 Thioredoxin_domain
IPR018253 DnaJ_domain_CS
IPR036249 Thioredoxin-like_sf
IPR001623 DnaJ_domain
IPR017937 Thioredoxin_CS
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR039755 TBC1D23
IPR000195 Rab-GTPase-TBC_dom
IPR036873 Rhodanese-like_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR001763 Rhodanese-like_dom
IPR013766 Thioredoxin_domain
IPR018253 DnaJ_domain_CS
IPR036249 Thioredoxin-like_sf
IPR001623 DnaJ_domain
IPR017937 Thioredoxin_CS
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR035979 RBD_domain_sf
IPR039755 TBC1D23
IPR000195 Rab-GTPase-TBC_dom
IPR036873 Rhodanese-like_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR001763 Rhodanese-like_dom
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H6VTR5
H9IZY6
A0A2H1WUX4
A0A194Q4A0
A0A212ENP8
A0A194QR87
+ More
A0A1S4F207 Q17I06 B0WYE9 A0A1Q3F4Q7 A0A1Q3F4T5 A0A1Q3F4V5 A0A182HAZ9 A0A1L8DTT7 A0A1B0F075 U5EYA9 W5JQP5 A0A084VX54 A0A182FJI2 A0A182J0H4 A0A336KHQ9 A0A182W631 A0A1I8NFP8 A0A1I8P9H9 F7ITY4 A0A067REI9 A0A2J7RJA7 A0A2J7RJA5 B4NA00 A0A0M4EMM9 A0A0L0C205 B4KYT8 B4J264 A0A154P8B5 B4LE75 E2AQZ9 A0A2P8YH34 A0A1Y1N5Y4 A0A2A3EIQ6 D6X059 A0A182PTY3 A0A088A8H8 A0A0A9YJ78 A0A0K8T8U9 A0A310S6U1 A0A0J7KXD5 A0A195EL35 A0A195FC91 A0A0C9QAM9 E2BWJ4 A0A182W7L1 A0A0C9QVY4 A0A195CJ56 A0A151WR59 A0A1B6DHU2 A0A0R3P1Z5 A0A1J1J813 A0A026VTH3 A0A158NZJ1 A0A195BMT1 T1HM40 A0A182YQS5 A0A182MHE4 A0A182RVV9 A0A1A9YUE9 K7J2H4 A0A3B0KT24 A0A232ESD4 A0A0K8VAP4 A0A1B6K467 A0A1B6GKN3 W8BJ44 A0A182RJD8 A0A0K8UY57 A0A1A9W3K0 A0A224X855 A0A0Q5WLP3 A0A0P8XDV4 A0A3L8D825 A0A0A1WZF6 A0A0K2VET6 Q7PLG1 E0VK40 A0A087URH1 A0A1S3DS69 A0A0P4VW10 A0A1A9XZD4 A0A0P4VY93 A0A1B0BJE7 V5GFC9 Q6AWH1 A0A1S4EDK0 A0A3B0KIN6 A0A293M034 A0A0P4XQW3 A0A0N8BGB1 A0A2P2I9R6 A0A0P4XYJ1 A0A0P5NML7 A0A0P5T3I9
A0A1S4F207 Q17I06 B0WYE9 A0A1Q3F4Q7 A0A1Q3F4T5 A0A1Q3F4V5 A0A182HAZ9 A0A1L8DTT7 A0A1B0F075 U5EYA9 W5JQP5 A0A084VX54 A0A182FJI2 A0A182J0H4 A0A336KHQ9 A0A182W631 A0A1I8NFP8 A0A1I8P9H9 F7ITY4 A0A067REI9 A0A2J7RJA7 A0A2J7RJA5 B4NA00 A0A0M4EMM9 A0A0L0C205 B4KYT8 B4J264 A0A154P8B5 B4LE75 E2AQZ9 A0A2P8YH34 A0A1Y1N5Y4 A0A2A3EIQ6 D6X059 A0A182PTY3 A0A088A8H8 A0A0A9YJ78 A0A0K8T8U9 A0A310S6U1 A0A0J7KXD5 A0A195EL35 A0A195FC91 A0A0C9QAM9 E2BWJ4 A0A182W7L1 A0A0C9QVY4 A0A195CJ56 A0A151WR59 A0A1B6DHU2 A0A0R3P1Z5 A0A1J1J813 A0A026VTH3 A0A158NZJ1 A0A195BMT1 T1HM40 A0A182YQS5 A0A182MHE4 A0A182RVV9 A0A1A9YUE9 K7J2H4 A0A3B0KT24 A0A232ESD4 A0A0K8VAP4 A0A1B6K467 A0A1B6GKN3 W8BJ44 A0A182RJD8 A0A0K8UY57 A0A1A9W3K0 A0A224X855 A0A0Q5WLP3 A0A0P8XDV4 A0A3L8D825 A0A0A1WZF6 A0A0K2VET6 Q7PLG1 E0VK40 A0A087URH1 A0A1S3DS69 A0A0P4VW10 A0A1A9XZD4 A0A0P4VY93 A0A1B0BJE7 V5GFC9 Q6AWH1 A0A1S4EDK0 A0A3B0KIN6 A0A293M034 A0A0P4XQW3 A0A0N8BGB1 A0A2P2I9R6 A0A0P4XYJ1 A0A0P5NML7 A0A0P5T3I9
PDB
2CUG
E-value=1.32176e-20,
Score=245
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.891555
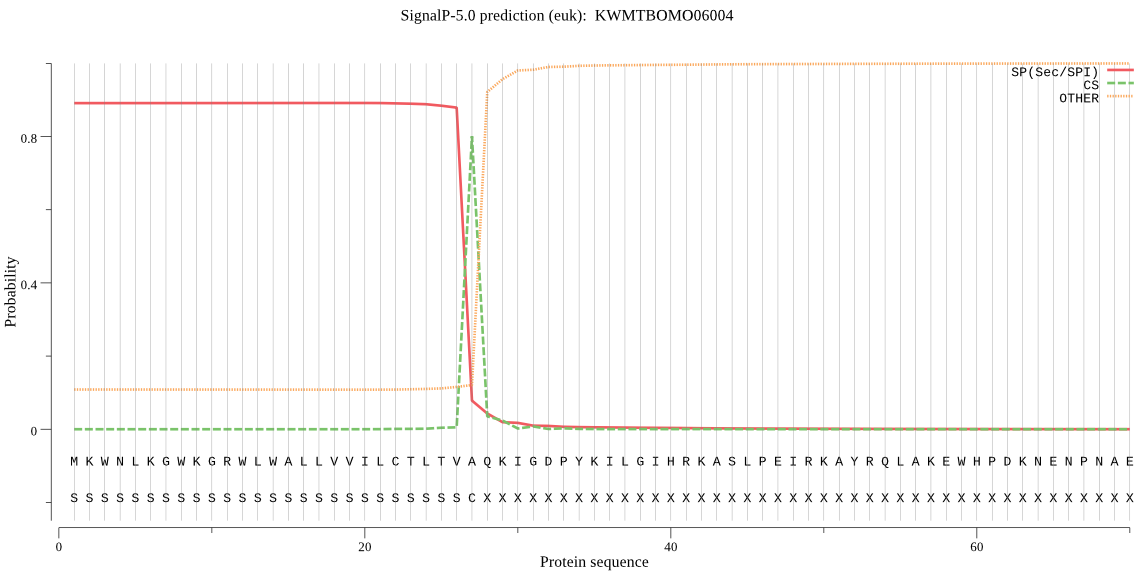
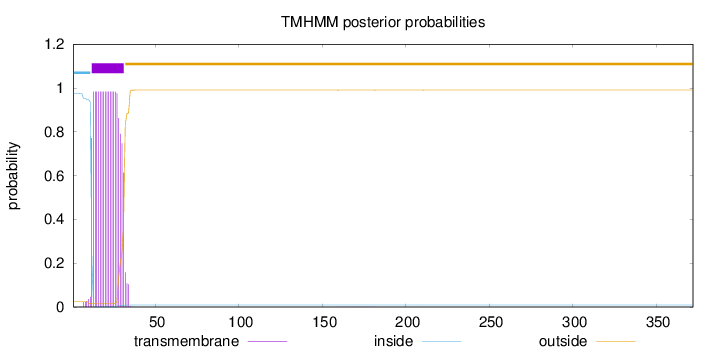
Length:
372
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.07056
Exp number, first 60 AAs:
19.06314
Total prob of N-in:
0.97518
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 31
outside
32 - 372
Population Genetic Test Statistics
Pi
22.159945
Theta
13.358794
Tajima's D
-2.000312
CLR
40.362941
CSRT
0.0131493425328734
Interpretation
Uncertain
