Gene
KWMTBOMO06003
Pre Gene Modal
BGIBMGA002881
Annotation
Sugar_transporter_SWEET1_[Papilio_machaon]
Full name
Sugar transporter SWEET
Location in the cell
PlasmaMembrane Reliability : 4.954
Sequence
CDS
ATGGACGCCTTAGCGGATATTCTTCAACCGTACAAAGAGATAGTGGGCTCGATAGCGGCCTTCGTAACCATTGGGCAGATGTTCTCCGGCAGTTTCATATGCTGGGACATATACAAGCAGGGCAGCACTCGCGGGATCGGCATCATGCCGTTCCTCGGAGGTGTTGTTATGACCATCCTCAACTTGCAATATGGTTACATACTCCGTGACGACACCATGATTCAAGTCAACATGTTCGGTCTGGCCTTAAATATCGTCTACGTGCTGATATACTTCAATTACACCCAGCAGAAGGTCAAGGTTTGGGCTCAGATCGGTCTGGCCGGTGCTGTGTCAGCTGCTCTTATAGGATACGCCCAGACCGAAGATCCTAAGCTCGTGGAGACCAGATTCGGGACAATAATAACGGCATTCATGTTCTACTTGATAGCTTCACCGCTTTTTGGACTGAAAGAGATCATTCAGAATAAGAGCACAGAAGGCATGCCGTTCCCTATAATACTGTCCGGAACTGTGGTCACGTTCATGTGGCTACTGTATGGTATTATATTGAGGAATAAATTCTTAGTGGTACAAAACGTGGTGGCCTTAGTGCTGTGCGCATCGCAACTGTCACTCTTCGTTATATACCCATCCAAGCCAAAAGGCAAAGATAAATCGAAAGTAAAAGCCAAGAAGACCGAGTAA
Protein
MDALADILQPYKEIVGSIAAFVTIGQMFSGSFICWDIYKQGSTRGIGIMPFLGGVVMTILNLQYGYILRDDTMIQVNMFGLALNIVYVLIYFNYTQQKVKVWAQIGLAGAVSAALIGYAQTEDPKLVETRFGTIITAFMFYLIASPLFGLKEIIQNKSTEGMPFPIILSGTVVTFMWLLYGIILRNKFLVVQNVVALVLCASQLSLFVIYPSKPKGKDKSKVKAKKTE
Summary
Description
Mediates sugar transport across membranes.
Similarity
Belongs to the SWEET sugar transporter family.
Uniprot
H9J046
A0A2A4JP00
A0A3S2NS34
A0A194QWQ2
A0A194Q3Q9
A0A1E1WNJ7
+ More
S4P1Q5 A0A212ENU7 A0A1B0CJB5 Q17A97 A0A182WCD7 A0A023EK24 A0A182LRW5 A0A1B0DA46 A0A1L8DDV4 A0A182YLE6 A0A2M4BZS6 A0A2M4AG31 Q7Q5N8 A0A182VL91 A0A182SNM7 W5JQI5 A0A2M3ZL04 A0A2M4AGT1 A0A182FC66 A0A182QZG2 A0A182J313 A0A182NKB3 A0A182IMR6 A0A1Q3FRT4 B0WXK1 A0A1Q3FRD3 A0A182GXU7 A0A023EMS5 Q174X0 A0A182P876 A0A182QQV9 A0A1Q3FRV8 A0A182TX56 A0A182LLA9 A0A182I0Z6 A0A336M9X7 A0A182X0U3 A0A084WT50 U5ESX2 A0A182XWL9 B0X578 A0A182UEC4 A0A182KWS9 A0A182V0T5 A0NDW0 A0A182VYA0 A0A0A8J811 A0A182MM18 A0A182NSS6 A0A182FZX7 A0A182PVH6 W5JGX7 A0A232FJR4 A0A1B6GRM6 A0A0N0BEJ1 A0A067RAA7 D6X0R8 A0A0L7RKC8 A0A0K8TRD9 A0A182KBG0 A0A2J7PML7 A0A2P8ZJT7 A0A1B6BYL0 J3JW37 A0A2A3E8V6 V9IFF2 A0A087ZRA4 A0A0C9RNF1 A0A023F841 U5ESW2 W8AR93 A0A182HJL9 A0A182RFY0 A0A1J1IW73 A0A0L7KXQ9 B4N501 A0A154PLV9 B4LH68 B4L148 A0A146KPY9 A0A2S2QF45 A0A1I8ML58 T1PC97 B3M713 A0A224XK00 E2A1M7 A0A1L8EFK3 B4IZ11 U4UVD4 A0A0M3QWF3 B4K1W1 A0A1Y1LRR4 A0A1I8PQR4
S4P1Q5 A0A212ENU7 A0A1B0CJB5 Q17A97 A0A182WCD7 A0A023EK24 A0A182LRW5 A0A1B0DA46 A0A1L8DDV4 A0A182YLE6 A0A2M4BZS6 A0A2M4AG31 Q7Q5N8 A0A182VL91 A0A182SNM7 W5JQI5 A0A2M3ZL04 A0A2M4AGT1 A0A182FC66 A0A182QZG2 A0A182J313 A0A182NKB3 A0A182IMR6 A0A1Q3FRT4 B0WXK1 A0A1Q3FRD3 A0A182GXU7 A0A023EMS5 Q174X0 A0A182P876 A0A182QQV9 A0A1Q3FRV8 A0A182TX56 A0A182LLA9 A0A182I0Z6 A0A336M9X7 A0A182X0U3 A0A084WT50 U5ESX2 A0A182XWL9 B0X578 A0A182UEC4 A0A182KWS9 A0A182V0T5 A0NDW0 A0A182VYA0 A0A0A8J811 A0A182MM18 A0A182NSS6 A0A182FZX7 A0A182PVH6 W5JGX7 A0A232FJR4 A0A1B6GRM6 A0A0N0BEJ1 A0A067RAA7 D6X0R8 A0A0L7RKC8 A0A0K8TRD9 A0A182KBG0 A0A2J7PML7 A0A2P8ZJT7 A0A1B6BYL0 J3JW37 A0A2A3E8V6 V9IFF2 A0A087ZRA4 A0A0C9RNF1 A0A023F841 U5ESW2 W8AR93 A0A182HJL9 A0A182RFY0 A0A1J1IW73 A0A0L7KXQ9 B4N501 A0A154PLV9 B4LH68 B4L148 A0A146KPY9 A0A2S2QF45 A0A1I8ML58 T1PC97 B3M713 A0A224XK00 E2A1M7 A0A1L8EFK3 B4IZ11 U4UVD4 A0A0M3QWF3 B4K1W1 A0A1Y1LRR4 A0A1I8PQR4
Pubmed
EMBL
BABH01018946
NWSH01000943
PCG73438.1
RSAL01000002
RVE54953.1
KQ461175
+ More
KPJ08001.1 KQ459472 KPJ00173.1 GDQN01002474 JAT88580.1 GAIX01010352 JAA82208.1 AGBW02013618 OWR43121.1 AJWK01014407 CH477337 EAT43194.1 GAPW01004138 JAC09460.1 AXCM01002455 AJVK01028821 GFDF01009580 JAV04504.1 GGFJ01009097 MBW58238.1 GGFK01006422 MBW39743.1 AAAB01008960 EAA10852.2 ADMH02000627 ETN65558.1 GGFM01008389 MBW29140.1 GGFK01006682 MBW40003.1 AXCN02000411 GFDL01004849 JAV30196.1 DS232168 EDS36531.1 GFDL01004844 JAV30201.1 JXUM01020897 KQ560585 KXJ81784.1 GAPW01003954 JAC09644.1 CH477404 EAT41663.1 AXCN02000300 GFDL01004716 JAV30329.1 APCN01000001 UFQT01000609 SSX25699.1 ATLV01026841 KE525419 KFB53394.1 GANO01003047 JAB56824.1 DS232369 EDS40724.1 AAAB01008904 EAU76798.1 AB901082 BAQ02382.1 AXCM01010640 ADMH02001529 ETN62165.1 NNAY01000120 OXU30780.1 GECZ01004709 JAS65060.1 KQ435827 KOX71961.1 KK852823 KDR15496.1 KQ971372 EFA09565.2 KQ414573 KOC71289.1 GDAI01000905 JAI16698.1 NEVH01023973 PNF17582.1 PYGN01000035 PSN56762.1 GEDC01030947 GEDC01021500 JAS06351.1 JAS15798.1 APGK01058964 BT127455 KB741292 AEE62417.1 ENN70356.1 KZ288322 PBC28140.1 JR043705 AEY59800.1 GBYB01015027 JAG84794.1 GBBI01001256 JAC17456.1 GANO01003045 JAB56826.1 GAMC01015190 GAMC01015187 JAB91365.1 APCN01002185 CVRI01000063 CRL04461.1 JTDY01004626 KOB67930.1 CH964101 EDW79440.1 KQ434943 KZC12210.1 CH940647 EDW70581.1 CH933809 EDW18205.1 GDHC01021292 GDHC01013521 GDHC01009579 GDHC01005334 GDHC01001545 JAP97336.1 JAQ05108.1 JAQ09050.1 JAQ13295.1 JAQ17084.1 GGMS01007155 MBY76358.1 KA645558 AFP60187.1 CH902618 EDV40878.1 GFTR01003610 JAW12816.1 GL435766 EFN72700.1 GFDG01001318 JAV17481.1 CH916366 EDV97719.1 KB632383 ERL94201.1 CP012525 ALC44049.1 CH917929 EDW04688.1 GEZM01054384 JAV73707.1
KPJ08001.1 KQ459472 KPJ00173.1 GDQN01002474 JAT88580.1 GAIX01010352 JAA82208.1 AGBW02013618 OWR43121.1 AJWK01014407 CH477337 EAT43194.1 GAPW01004138 JAC09460.1 AXCM01002455 AJVK01028821 GFDF01009580 JAV04504.1 GGFJ01009097 MBW58238.1 GGFK01006422 MBW39743.1 AAAB01008960 EAA10852.2 ADMH02000627 ETN65558.1 GGFM01008389 MBW29140.1 GGFK01006682 MBW40003.1 AXCN02000411 GFDL01004849 JAV30196.1 DS232168 EDS36531.1 GFDL01004844 JAV30201.1 JXUM01020897 KQ560585 KXJ81784.1 GAPW01003954 JAC09644.1 CH477404 EAT41663.1 AXCN02000300 GFDL01004716 JAV30329.1 APCN01000001 UFQT01000609 SSX25699.1 ATLV01026841 KE525419 KFB53394.1 GANO01003047 JAB56824.1 DS232369 EDS40724.1 AAAB01008904 EAU76798.1 AB901082 BAQ02382.1 AXCM01010640 ADMH02001529 ETN62165.1 NNAY01000120 OXU30780.1 GECZ01004709 JAS65060.1 KQ435827 KOX71961.1 KK852823 KDR15496.1 KQ971372 EFA09565.2 KQ414573 KOC71289.1 GDAI01000905 JAI16698.1 NEVH01023973 PNF17582.1 PYGN01000035 PSN56762.1 GEDC01030947 GEDC01021500 JAS06351.1 JAS15798.1 APGK01058964 BT127455 KB741292 AEE62417.1 ENN70356.1 KZ288322 PBC28140.1 JR043705 AEY59800.1 GBYB01015027 JAG84794.1 GBBI01001256 JAC17456.1 GANO01003045 JAB56826.1 GAMC01015190 GAMC01015187 JAB91365.1 APCN01002185 CVRI01000063 CRL04461.1 JTDY01004626 KOB67930.1 CH964101 EDW79440.1 KQ434943 KZC12210.1 CH940647 EDW70581.1 CH933809 EDW18205.1 GDHC01021292 GDHC01013521 GDHC01009579 GDHC01005334 GDHC01001545 JAP97336.1 JAQ05108.1 JAQ09050.1 JAQ13295.1 JAQ17084.1 GGMS01007155 MBY76358.1 KA645558 AFP60187.1 CH902618 EDV40878.1 GFTR01003610 JAW12816.1 GL435766 EFN72700.1 GFDG01001318 JAV17481.1 CH916366 EDV97719.1 KB632383 ERL94201.1 CP012525 ALC44049.1 CH917929 EDW04688.1 GEZM01054384 JAV73707.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000092461 UP000008820 UP000075920 UP000075883 UP000092462 UP000076408 UP000007062 UP000075903 UP000075901 UP000000673 UP000069272 UP000075886 UP000075880 UP000075884 UP000002320 UP000069940 UP000249989 UP000075885 UP000075902 UP000075882 UP000075840 UP000076407 UP000030765 UP000215335 UP000053105 UP000027135 UP000007266 UP000053825 UP000075881 UP000235965 UP000245037 UP000019118 UP000242457 UP000005203 UP000075900 UP000183832 UP000037510 UP000007798 UP000076502 UP000008792 UP000009192 UP000095301 UP000007801 UP000000311 UP000001070 UP000030742 UP000092553 UP000095300
UP000092461 UP000008820 UP000075920 UP000075883 UP000092462 UP000076408 UP000007062 UP000075903 UP000075901 UP000000673 UP000069272 UP000075886 UP000075880 UP000075884 UP000002320 UP000069940 UP000249989 UP000075885 UP000075902 UP000075882 UP000075840 UP000076407 UP000030765 UP000215335 UP000053105 UP000027135 UP000007266 UP000053825 UP000075881 UP000235965 UP000245037 UP000019118 UP000242457 UP000005203 UP000075900 UP000183832 UP000037510 UP000007798 UP000076502 UP000008792 UP000009192 UP000095301 UP000007801 UP000000311 UP000001070 UP000030742 UP000092553 UP000095300
PRIDE
Pfam
PF03083 MtN3_slv
Interpro
IPR004316
SWEET_sugar_transpr
ProteinModelPortal
H9J046
A0A2A4JP00
A0A3S2NS34
A0A194QWQ2
A0A194Q3Q9
A0A1E1WNJ7
+ More
S4P1Q5 A0A212ENU7 A0A1B0CJB5 Q17A97 A0A182WCD7 A0A023EK24 A0A182LRW5 A0A1B0DA46 A0A1L8DDV4 A0A182YLE6 A0A2M4BZS6 A0A2M4AG31 Q7Q5N8 A0A182VL91 A0A182SNM7 W5JQI5 A0A2M3ZL04 A0A2M4AGT1 A0A182FC66 A0A182QZG2 A0A182J313 A0A182NKB3 A0A182IMR6 A0A1Q3FRT4 B0WXK1 A0A1Q3FRD3 A0A182GXU7 A0A023EMS5 Q174X0 A0A182P876 A0A182QQV9 A0A1Q3FRV8 A0A182TX56 A0A182LLA9 A0A182I0Z6 A0A336M9X7 A0A182X0U3 A0A084WT50 U5ESX2 A0A182XWL9 B0X578 A0A182UEC4 A0A182KWS9 A0A182V0T5 A0NDW0 A0A182VYA0 A0A0A8J811 A0A182MM18 A0A182NSS6 A0A182FZX7 A0A182PVH6 W5JGX7 A0A232FJR4 A0A1B6GRM6 A0A0N0BEJ1 A0A067RAA7 D6X0R8 A0A0L7RKC8 A0A0K8TRD9 A0A182KBG0 A0A2J7PML7 A0A2P8ZJT7 A0A1B6BYL0 J3JW37 A0A2A3E8V6 V9IFF2 A0A087ZRA4 A0A0C9RNF1 A0A023F841 U5ESW2 W8AR93 A0A182HJL9 A0A182RFY0 A0A1J1IW73 A0A0L7KXQ9 B4N501 A0A154PLV9 B4LH68 B4L148 A0A146KPY9 A0A2S2QF45 A0A1I8ML58 T1PC97 B3M713 A0A224XK00 E2A1M7 A0A1L8EFK3 B4IZ11 U4UVD4 A0A0M3QWF3 B4K1W1 A0A1Y1LRR4 A0A1I8PQR4
S4P1Q5 A0A212ENU7 A0A1B0CJB5 Q17A97 A0A182WCD7 A0A023EK24 A0A182LRW5 A0A1B0DA46 A0A1L8DDV4 A0A182YLE6 A0A2M4BZS6 A0A2M4AG31 Q7Q5N8 A0A182VL91 A0A182SNM7 W5JQI5 A0A2M3ZL04 A0A2M4AGT1 A0A182FC66 A0A182QZG2 A0A182J313 A0A182NKB3 A0A182IMR6 A0A1Q3FRT4 B0WXK1 A0A1Q3FRD3 A0A182GXU7 A0A023EMS5 Q174X0 A0A182P876 A0A182QQV9 A0A1Q3FRV8 A0A182TX56 A0A182LLA9 A0A182I0Z6 A0A336M9X7 A0A182X0U3 A0A084WT50 U5ESX2 A0A182XWL9 B0X578 A0A182UEC4 A0A182KWS9 A0A182V0T5 A0NDW0 A0A182VYA0 A0A0A8J811 A0A182MM18 A0A182NSS6 A0A182FZX7 A0A182PVH6 W5JGX7 A0A232FJR4 A0A1B6GRM6 A0A0N0BEJ1 A0A067RAA7 D6X0R8 A0A0L7RKC8 A0A0K8TRD9 A0A182KBG0 A0A2J7PML7 A0A2P8ZJT7 A0A1B6BYL0 J3JW37 A0A2A3E8V6 V9IFF2 A0A087ZRA4 A0A0C9RNF1 A0A023F841 U5ESW2 W8AR93 A0A182HJL9 A0A182RFY0 A0A1J1IW73 A0A0L7KXQ9 B4N501 A0A154PLV9 B4LH68 B4L148 A0A146KPY9 A0A2S2QF45 A0A1I8ML58 T1PC97 B3M713 A0A224XK00 E2A1M7 A0A1L8EFK3 B4IZ11 U4UVD4 A0A0M3QWF3 B4K1W1 A0A1Y1LRR4 A0A1I8PQR4
PDB
5XPD
E-value=6.55011e-12,
Score=168
Ontologies
KEGG
GO
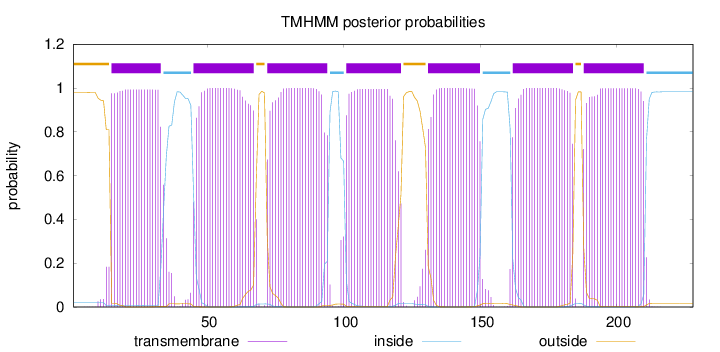
Topology
Subcellular location
Cell membrane
Length:
228
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.39347
Exp number, first 60 AAs:
35.70601
Total prob of N-in:
0.02052
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 33
inside
34 - 44
TMhelix
45 - 67
outside
68 - 71
TMhelix
72 - 94
inside
95 - 100
TMhelix
101 - 121
outside
122 - 130
TMhelix
131 - 150
inside
151 - 161
TMhelix
162 - 184
outside
185 - 187
TMhelix
188 - 210
inside
211 - 228
Population Genetic Test Statistics
Pi
17.992533
Theta
18.283936
Tajima's D
0.165834
CLR
0.975374
CSRT
0.420628968551572
Interpretation
Uncertain
