Gene
KWMTBOMO06001
Pre Gene Modal
BGIBMGA002883
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_2-oxoglutarate_dehydrogenase?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.333
Sequence
CDS
ATGCACGGAGACGCAGCGTTCTCTGGCCAGGGCGTTGTGTACGAGACAATGCATCTTGGAAACCTTCCGGCTTTTACCACACACGGTTCAATACACATTGTCTGCAACAACCAGATCGGGTACACGACGGATCCTAGGTTCGCGAGGAGCTCGCCTTACTGCTCTGACGTGGCGAAGTGTGTGGATGCTCCCGTGTTCCACGTGAACGCTGATGACGCGGAGGCCGTGGCGCACGTCGCGAGGGTCGCCATCGAGTACCGCTGCAAGTTCAAGAAGGACGTCGTGCTGGATCTGGTCTGCTATAGGCGCTTCGGGCACAGCGAAGAAGACGAGCCCATGTTCACTCAACCTTTCATGTACAAGAAAATTAAGAGCATGCAAACTGTTGATAAAATCTATGGCGAGAAACTGAAGGCAGAAGGTATAGTGACGGATGCCGATGTCAAGGGTTGGGAGAAAGAGTACATGGACACGCTAAATCGGCACTTTGAGCTCGCCAAGAAGGTCACCAAACTAAGCATTATGGATTGGATCGATACTCCATGGACTGGTTTCTTTGAGGCCAGAGATCCTAAGAAAGTAAGGTTTCAATAA
Protein
MHGDAAFSGQGVVYETMHLGNLPAFTTHGSIHIVCNNQIGYTTDPRFARSSPYCSDVAKCVDAPVFHVNADDAEAVAHVARVAIEYRCKFKKDVVLDLVCYRRFGHSEEDEPMFTQPFMYKKIKSMQTVDKIYGEKLKAEGIVTDADVKGWEKEYMDTLNRHFELAKKVTKLSIMDWIDTPWTGFFEARDPKKVRFQ
Summary
Uniprot
A0A212F1D6
A0A194QSN8
A0A194Q3R0
A0A2A4J669
A0A2A4JMC2
A0A2A4JNT4
+ More
A0A2H1WNX1 A0A0K8TMF4 A0A0T6BHJ8 A0A0A1X7U1 A0A1Y1LYV4 A0A1Y1LXZ0 V5SK40 A0A034WDZ4 A0A034WDE9 A0A0K8UFH9 A0A0A1WLY7 A0A0K8WD79 A0A034W9X1 A0A034WF05 A0A0K8VZ20 A0A034WCE3 A0A0A1XDZ0 W8C7N9 A0A0K8VW59 A0A0A1XMA9 W8BIT2 W8BZB1 W8BPJ7 A0A3R7MFH7 A0A1I8NP15 A0A1L8E1Z8 A0A1I8MZV3 A0A182GH27 A0A1L8E250 T1PDI6 Q8IGI6 A0A1B0B977 A0A1B0GI66 A0A1A9W7H8 A0A2H1VZY9 A0A1A9VJ75 A0A1A9XV32 A0A023EWR1 A0A1B0G1X4 Q175A3 Q175A4 T1GDU5 A0A0L0CD13 A0A1B0AEU1 B4IYB9 A0A0R1DYI5 A0A0B7B521 A0A0J9RWU4 A0A182G6V7 A0A0Q5UJL5 A0A0R1DY95 A0A1W4W5P2 A0A3B0JSY5 A0A0J9RWV3 A0A3B0JVW1 A0A0R3P923 A0A0J9RWU8 A0A0J9RWY6 A0A0Q9WW13 A0A0Q9WQS9 A0A0Q5U4S4 A0A0R3P8U0 Q29DU3 A0A3B0JSW0 A0A0R3P6Q4 A0A1W4VUH3 A0A0R3P6M5 A0A1W4VUI9 A0A0Q5U530 Q8IQQ0 A0A0J9RX36 A0A1W4W5N7 A0A0Q9WU89 Q9VVC5 A0A1W4W722 A0A0J9UMV3 A0A0Q9XCB5 A0A0Q9XQF1 A0A0R1E427 B4QND2 B4HK94 B4PK01 A0A0J9UMU8 A8JNU6 B3NDF1 B4LHN9 B4KW84 A0A0Q9XCC4 A0A336L6L3 A0A336L5D2 A0A226ESN0 A0A0Q9WW68 A0A0P8ZVE0 A0A0Q9WP13
A0A2H1WNX1 A0A0K8TMF4 A0A0T6BHJ8 A0A0A1X7U1 A0A1Y1LYV4 A0A1Y1LXZ0 V5SK40 A0A034WDZ4 A0A034WDE9 A0A0K8UFH9 A0A0A1WLY7 A0A0K8WD79 A0A034W9X1 A0A034WF05 A0A0K8VZ20 A0A034WCE3 A0A0A1XDZ0 W8C7N9 A0A0K8VW59 A0A0A1XMA9 W8BIT2 W8BZB1 W8BPJ7 A0A3R7MFH7 A0A1I8NP15 A0A1L8E1Z8 A0A1I8MZV3 A0A182GH27 A0A1L8E250 T1PDI6 Q8IGI6 A0A1B0B977 A0A1B0GI66 A0A1A9W7H8 A0A2H1VZY9 A0A1A9VJ75 A0A1A9XV32 A0A023EWR1 A0A1B0G1X4 Q175A3 Q175A4 T1GDU5 A0A0L0CD13 A0A1B0AEU1 B4IYB9 A0A0R1DYI5 A0A0B7B521 A0A0J9RWU4 A0A182G6V7 A0A0Q5UJL5 A0A0R1DY95 A0A1W4W5P2 A0A3B0JSY5 A0A0J9RWV3 A0A3B0JVW1 A0A0R3P923 A0A0J9RWU8 A0A0J9RWY6 A0A0Q9WW13 A0A0Q9WQS9 A0A0Q5U4S4 A0A0R3P8U0 Q29DU3 A0A3B0JSW0 A0A0R3P6Q4 A0A1W4VUH3 A0A0R3P6M5 A0A1W4VUI9 A0A0Q5U530 Q8IQQ0 A0A0J9RX36 A0A1W4W5N7 A0A0Q9WU89 Q9VVC5 A0A1W4W722 A0A0J9UMV3 A0A0Q9XCB5 A0A0Q9XQF1 A0A0R1E427 B4QND2 B4HK94 B4PK01 A0A0J9UMU8 A8JNU6 B3NDF1 B4LHN9 B4KW84 A0A0Q9XCC4 A0A336L6L3 A0A336L5D2 A0A226ESN0 A0A0Q9WW68 A0A0P8ZVE0 A0A0Q9WP13
Pubmed
EMBL
AGBW02010898
OWR47555.1
KQ461175
KPJ08000.1
KQ459472
KPJ00172.1
+ More
NWSH01003119 PCG66912.1 NWSH01001058 PCG72868.1 NWSH01000943 PCG73436.1 ODYU01009993 SOQ54773.1 GDAI01002277 JAI15326.1 LJIG01000119 KRT86821.1 GBXI01007316 JAD06976.1 GEZM01046472 GEZM01046471 GEZM01046470 GEZM01046468 GEZM01046466 JAV77215.1 GEZM01046467 GEZM01046465 JAV77220.1 KF647632 AHB50494.1 GAKP01006592 JAC52360.1 GAKP01006590 JAC52362.1 GDHF01026991 JAI25323.1 GBXI01014747 JAC99544.1 GDHF01005182 GDHF01003198 JAI47132.1 JAI49116.1 GAKP01006591 JAC52361.1 GAKP01006589 JAC52363.1 GDHF01020327 GDHF01008192 GDHF01002123 JAI31987.1 JAI44122.1 JAI50191.1 GAKP01006593 JAC52359.1 GBXI01005110 JAD09182.1 GAMC01007906 JAB98649.1 GDHF01009554 JAI42760.1 GBXI01002589 JAD11703.1 GAMC01007908 JAB98647.1 GAMC01007909 JAB98646.1 GAMC01007907 JAB98648.1 QCYY01001012 ROT81148.1 GFDF01001346 JAV12738.1 JXUM01011039 JXUM01011040 JXUM01011041 JXUM01011042 GFDF01001330 JAV12754.1 KA646852 AFP61481.1 BT001767 AAN71522.1 JXJN01010297 AJWK01011399 AJWK01011400 AJWK01011401 AJWK01011402 AJWK01011403 ODYU01005456 SOQ46333.1 GAPW01000081 JAC13517.1 CCAG010004394 CH477404 EAT41628.1 EAT41627.1 CAQQ02114576 CAQQ02114577 CAQQ02114578 JRES01000577 KNC30102.1 CH916366 EDV96569.1 CM000159 KRK02041.1 KRK02042.1 HACG01040525 HACG01040526 CEK87390.1 CEK87391.1 CM002912 KMZ00159.1 JXUM01045696 KQ561447 KXJ78560.1 CH954178 KQS44071.1 KQS44073.1 KQS44074.1 KRK02043.1 KRK02044.1 KRK02045.1 OUUW01000009 SPP85205.1 KMZ00158.1 KMZ00169.1 SPP85203.1 CH379070 KRT08819.1 KMZ00160.1 KMZ00164.1 KMZ00167.1 KMZ00162.1 CH940647 KRF84469.1 KRF84471.1 KQS44070.1 KQS44072.1 KRT08821.1 EAL30321.2 KRT08820.1 SPP85204.1 KRT08817.1 KRT08818.1 KQS44069.1 AE014296 BT012466 AAN11721.1 AAO41241.2 AAS93737.1 KMZ00163.1 KRF84468.1 KRF84470.1 BT003743 AAF49388.2 AAF49389.2 AAN11722.1 AAO41240.1 AAO41404.1 KMZ00165.1 KMZ00166.1 CH933809 KRG06207.1 KRG06211.1 KRK02039.1 CM000363 EDX10802.1 KMZ00156.1 CH480815 EDW41835.1 EDW94770.1 KRK02040.1 KMZ00157.1 KMZ00161.1 AAN11723.3 ABW08558.1 EDV52013.1 KQS44068.1 EDW69592.1 EDW18491.1 KRG06209.1 KRG06210.1 KRG06208.1 UFQS01002041 UFQT01002041 SSX13049.1 SSX32489.1 SSX13048.1 SSX32488.1 LNIX01000002 OXA60228.1 CH963847 KRF97620.1 CH902618 KPU78554.1 KRF97621.1
NWSH01003119 PCG66912.1 NWSH01001058 PCG72868.1 NWSH01000943 PCG73436.1 ODYU01009993 SOQ54773.1 GDAI01002277 JAI15326.1 LJIG01000119 KRT86821.1 GBXI01007316 JAD06976.1 GEZM01046472 GEZM01046471 GEZM01046470 GEZM01046468 GEZM01046466 JAV77215.1 GEZM01046467 GEZM01046465 JAV77220.1 KF647632 AHB50494.1 GAKP01006592 JAC52360.1 GAKP01006590 JAC52362.1 GDHF01026991 JAI25323.1 GBXI01014747 JAC99544.1 GDHF01005182 GDHF01003198 JAI47132.1 JAI49116.1 GAKP01006591 JAC52361.1 GAKP01006589 JAC52363.1 GDHF01020327 GDHF01008192 GDHF01002123 JAI31987.1 JAI44122.1 JAI50191.1 GAKP01006593 JAC52359.1 GBXI01005110 JAD09182.1 GAMC01007906 JAB98649.1 GDHF01009554 JAI42760.1 GBXI01002589 JAD11703.1 GAMC01007908 JAB98647.1 GAMC01007909 JAB98646.1 GAMC01007907 JAB98648.1 QCYY01001012 ROT81148.1 GFDF01001346 JAV12738.1 JXUM01011039 JXUM01011040 JXUM01011041 JXUM01011042 GFDF01001330 JAV12754.1 KA646852 AFP61481.1 BT001767 AAN71522.1 JXJN01010297 AJWK01011399 AJWK01011400 AJWK01011401 AJWK01011402 AJWK01011403 ODYU01005456 SOQ46333.1 GAPW01000081 JAC13517.1 CCAG010004394 CH477404 EAT41628.1 EAT41627.1 CAQQ02114576 CAQQ02114577 CAQQ02114578 JRES01000577 KNC30102.1 CH916366 EDV96569.1 CM000159 KRK02041.1 KRK02042.1 HACG01040525 HACG01040526 CEK87390.1 CEK87391.1 CM002912 KMZ00159.1 JXUM01045696 KQ561447 KXJ78560.1 CH954178 KQS44071.1 KQS44073.1 KQS44074.1 KRK02043.1 KRK02044.1 KRK02045.1 OUUW01000009 SPP85205.1 KMZ00158.1 KMZ00169.1 SPP85203.1 CH379070 KRT08819.1 KMZ00160.1 KMZ00164.1 KMZ00167.1 KMZ00162.1 CH940647 KRF84469.1 KRF84471.1 KQS44070.1 KQS44072.1 KRT08821.1 EAL30321.2 KRT08820.1 SPP85204.1 KRT08817.1 KRT08818.1 KQS44069.1 AE014296 BT012466 AAN11721.1 AAO41241.2 AAS93737.1 KMZ00163.1 KRF84468.1 KRF84470.1 BT003743 AAF49388.2 AAF49389.2 AAN11722.1 AAO41240.1 AAO41404.1 KMZ00165.1 KMZ00166.1 CH933809 KRG06207.1 KRG06211.1 KRK02039.1 CM000363 EDX10802.1 KMZ00156.1 CH480815 EDW41835.1 EDW94770.1 KRK02040.1 KMZ00157.1 KMZ00161.1 AAN11723.3 ABW08558.1 EDV52013.1 KQS44068.1 EDW69592.1 EDW18491.1 KRG06209.1 KRG06210.1 KRG06208.1 UFQS01002041 UFQT01002041 SSX13049.1 SSX32489.1 SSX13048.1 SSX32488.1 LNIX01000002 OXA60228.1 CH963847 KRF97620.1 CH902618 KPU78554.1 KRF97621.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000283509
UP000095300
+ More
UP000095301 UP000069940 UP000092460 UP000092461 UP000091820 UP000078200 UP000092443 UP000092444 UP000008820 UP000015102 UP000037069 UP000092445 UP000001070 UP000002282 UP000249989 UP000008711 UP000192221 UP000268350 UP000001819 UP000008792 UP000000803 UP000009192 UP000000304 UP000001292 UP000198287 UP000007798 UP000007801
UP000095301 UP000069940 UP000092460 UP000092461 UP000091820 UP000078200 UP000092443 UP000092444 UP000008820 UP000015102 UP000037069 UP000092445 UP000001070 UP000002282 UP000249989 UP000008711 UP000192221 UP000268350 UP000001819 UP000008792 UP000000803 UP000009192 UP000000304 UP000001292 UP000198287 UP000007798 UP000007801
Interpro
SUPFAM
SSF52518
SSF52518
Gene 3D
ProteinModelPortal
A0A212F1D6
A0A194QSN8
A0A194Q3R0
A0A2A4J669
A0A2A4JMC2
A0A2A4JNT4
+ More
A0A2H1WNX1 A0A0K8TMF4 A0A0T6BHJ8 A0A0A1X7U1 A0A1Y1LYV4 A0A1Y1LXZ0 V5SK40 A0A034WDZ4 A0A034WDE9 A0A0K8UFH9 A0A0A1WLY7 A0A0K8WD79 A0A034W9X1 A0A034WF05 A0A0K8VZ20 A0A034WCE3 A0A0A1XDZ0 W8C7N9 A0A0K8VW59 A0A0A1XMA9 W8BIT2 W8BZB1 W8BPJ7 A0A3R7MFH7 A0A1I8NP15 A0A1L8E1Z8 A0A1I8MZV3 A0A182GH27 A0A1L8E250 T1PDI6 Q8IGI6 A0A1B0B977 A0A1B0GI66 A0A1A9W7H8 A0A2H1VZY9 A0A1A9VJ75 A0A1A9XV32 A0A023EWR1 A0A1B0G1X4 Q175A3 Q175A4 T1GDU5 A0A0L0CD13 A0A1B0AEU1 B4IYB9 A0A0R1DYI5 A0A0B7B521 A0A0J9RWU4 A0A182G6V7 A0A0Q5UJL5 A0A0R1DY95 A0A1W4W5P2 A0A3B0JSY5 A0A0J9RWV3 A0A3B0JVW1 A0A0R3P923 A0A0J9RWU8 A0A0J9RWY6 A0A0Q9WW13 A0A0Q9WQS9 A0A0Q5U4S4 A0A0R3P8U0 Q29DU3 A0A3B0JSW0 A0A0R3P6Q4 A0A1W4VUH3 A0A0R3P6M5 A0A1W4VUI9 A0A0Q5U530 Q8IQQ0 A0A0J9RX36 A0A1W4W5N7 A0A0Q9WU89 Q9VVC5 A0A1W4W722 A0A0J9UMV3 A0A0Q9XCB5 A0A0Q9XQF1 A0A0R1E427 B4QND2 B4HK94 B4PK01 A0A0J9UMU8 A8JNU6 B3NDF1 B4LHN9 B4KW84 A0A0Q9XCC4 A0A336L6L3 A0A336L5D2 A0A226ESN0 A0A0Q9WW68 A0A0P8ZVE0 A0A0Q9WP13
A0A2H1WNX1 A0A0K8TMF4 A0A0T6BHJ8 A0A0A1X7U1 A0A1Y1LYV4 A0A1Y1LXZ0 V5SK40 A0A034WDZ4 A0A034WDE9 A0A0K8UFH9 A0A0A1WLY7 A0A0K8WD79 A0A034W9X1 A0A034WF05 A0A0K8VZ20 A0A034WCE3 A0A0A1XDZ0 W8C7N9 A0A0K8VW59 A0A0A1XMA9 W8BIT2 W8BZB1 W8BPJ7 A0A3R7MFH7 A0A1I8NP15 A0A1L8E1Z8 A0A1I8MZV3 A0A182GH27 A0A1L8E250 T1PDI6 Q8IGI6 A0A1B0B977 A0A1B0GI66 A0A1A9W7H8 A0A2H1VZY9 A0A1A9VJ75 A0A1A9XV32 A0A023EWR1 A0A1B0G1X4 Q175A3 Q175A4 T1GDU5 A0A0L0CD13 A0A1B0AEU1 B4IYB9 A0A0R1DYI5 A0A0B7B521 A0A0J9RWU4 A0A182G6V7 A0A0Q5UJL5 A0A0R1DY95 A0A1W4W5P2 A0A3B0JSY5 A0A0J9RWV3 A0A3B0JVW1 A0A0R3P923 A0A0J9RWU8 A0A0J9RWY6 A0A0Q9WW13 A0A0Q9WQS9 A0A0Q5U4S4 A0A0R3P8U0 Q29DU3 A0A3B0JSW0 A0A0R3P6Q4 A0A1W4VUH3 A0A0R3P6M5 A0A1W4VUI9 A0A0Q5U530 Q8IQQ0 A0A0J9RX36 A0A1W4W5N7 A0A0Q9WU89 Q9VVC5 A0A1W4W722 A0A0J9UMV3 A0A0Q9XCB5 A0A0Q9XQF1 A0A0R1E427 B4QND2 B4HK94 B4PK01 A0A0J9UMU8 A8JNU6 B3NDF1 B4LHN9 B4KW84 A0A0Q9XCC4 A0A336L6L3 A0A336L5D2 A0A226ESN0 A0A0Q9WW68 A0A0P8ZVE0 A0A0Q9WP13
PDB
2XT6
E-value=1.86156e-46,
Score=465
Ontologies
GO
PANTHER
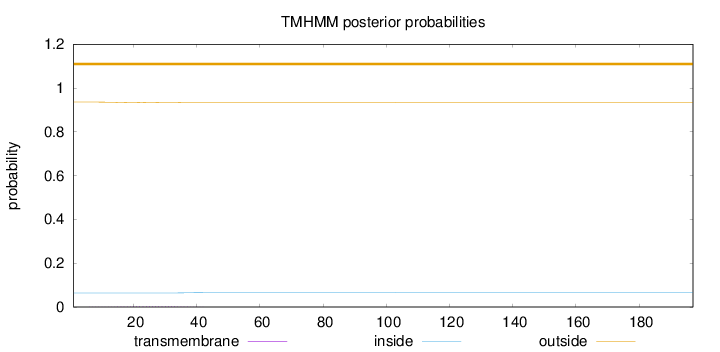
Topology
Length:
197
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0633800000000001
Exp number, first 60 AAs:
0.06304
Total prob of N-in:
0.06418
outside
1 - 197
Population Genetic Test Statistics
Pi
14.832011
Theta
15.845288
Tajima's D
-0.192665
CLR
3.948146
CSRT
0.321633918304085
Interpretation
Uncertain
