Gene
KWMTBOMO05999 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002820
Annotation
PREDICTED:_transmembrane_GTPase_Marf_isoform_X3_[Amyelois_transitella]
Full name
Transmembrane GTPase Marf
Alternative Name
Mitochondrial assembly regulatory factor
Mitofusin
Mitofusin
Location in the cell
Cytoplasmic Reliability : 1.393 Mitochondrial Reliability : 1.103 Nuclear Reliability : 1.593
Sequence
CDS
ATGGCGGCGTATGTTAACCGAACGATATCGATGATGGCTGGTGACGGGCCACACAATGTGTCGATGTTGAACAACGGCTCGGTACGTGTGAACATGCAAAATGTTGACTCCCCGCTCCAGATATTCGTCAGAGCGAAGAAGAAGATAAACGATATATTTGTGGAGATTGACGATTATGTCAAAGATGCTGTCACTTTTATGCATGCTGTATCGGGTGAGAATGGCATTGCCACACCACAGGATATGGGCAATGTGGAGTCATATGTGTCAAAGGTGGAAGCCATCAGAGAGGTTCTCAAGCGAGATCATATGAAAGTTGCTTTCTTTGGAAGGACAAGCAATGGCAAAAGCACAGTTATAAATGCGATGCTCCATGACAAAATCCTACCGAGCGGGATAGGACACACGACTAACTGCTTCTTGCAAGTTGAAGGTTCTGATACAAACGAAGCCTACATGAGAACGGAAGGCTGCGAGGAGAAACTTAATGTACAGTCCGTAAGCCAATTAGGTCACGCCCTGTGCGCGACTCGTCTGCAAGAGTGCTCGCTGGTGCACGTGCACTGGCCCCGCGAGCTGTGCGCGCTGCTGCGGGACGACGTGGTGCTGGTGGACAGCCCCGGCGTGGACGTGACGCCGAACCTCGACACGTGGATCGACAAGTACTGCCTCGACGCGGACGTGTTCGTGCTCGTGGCCAACGCAGAGTCCACGCTCATGGTCACTGAGAAGAACTTCTTTCACAAAGTATCGACGAAGATATCCCAACCGAATATATTTATACTGAACAATCGCTGGGACGCGTCCGCCTCCGAGCCGGAGTATATGGAGCAGGTACGCACACAACACGCGAACCGCTGCGTCGACTTCTTATCGCGTGAGCTGCGTGTGTGCTCCCCTAAAGAGGCTGAGGAGAGGATCTTCTTCATATCCGCCAAGGAAGCCCTGCTCACCAGGATGCGTGATCGAGAGAAACCTGTCTCTTCTCCGATCCTGGCGGAGGGTCACCAGGTGCGCTACTTCGAGTTCGTGGACTTCGAGCGCAAGTTTGAGGAGTGCATCAGCCAGTCGGCCGTGCGCACCAAGTTCGCGCAGCACTCGCGCCGCGGCAAGAACATCGCTGGGGACGTCATGGCGGCTCTCGACCGGGTGTACAACATAGCAACAGAGCAAAAGGCAGCCAAGGTCGAGAAGCAGAGGATACTGCACGAGCAGCTGTCGTCCATAGAGGAACAGTTGACCACCATCACGAGGCAGATGAAGGACAAGATCAACAGGATGGTTGAGAGCGTGGAGCACAAGGTATCTCTAACCCTGAGTCAAGAGATCCGTCGTCTGTCGGCCCTCGTCGATGAATACGAATCGGAATTCCGTCCGGAGCGACCCGCCCTCGAGCAGTACAAGCGAGCCCTGCACCGGCACGTCGAGGCGGGGCTAGGGTCTAGACTCAAGAAGAGACTATCTTCAGATATAGGAAACGAAATGGACGTAGTTCAAAAGGAGATGGCCGAACGCATGTACAACATCCTCCCCACCAACAAGCGGGCCGCCGCAGCGAACTACATCATACCTCACCAGCAGCCCTTCGAGGTGCTGTACCGGCTGAACTGCGACAACCTCTGCGCCGACTTCAACGAAGACTTGTCGTTCCGCTTCTCGTACGGGATCACGGCTCTCATCCAACGGTTCCAGGGAAAGAATACTAATCGGATCGCTTTGAATAACCCCCCACAGTACACTCAGATCCCGGCCAGCATCACCCCGGTGTCGGCGTTGCCCCCCGCCCCCCGCGCCGCGCTGCCCGTGCTCAGCGCGGAGGAGTGGGGGCTGCTCTCCAAGTTCGCGCTGGGGGCCCTCACGTCGCAGGGGACCATGGGGGGCCTGTTGCTCTCCGGATTGCTGCTGAAGACGGTGGGGTGGCGCGTGCTGGCGGTGACGGGGCTGGCGTACGGCGCGCTGTACGCGTACGAGCGCCTCACGTGGACGGCCAAGGCGCAGGAGCGCGTCTTCAAGCGCCAGTACGTCGCGCACGCCGGCAAGAAGCTGCGCCTCATCGTCGACCTCACCTCCGCCAACTGCAGCCACCAAGTGCAGCAGGAGCTGTCCACGATGTTCGCCCGGCTGTGTCGGCTCATCGACGAGGCCACCACGGAGATGGACGCCGAGCTGTCCGGGGTGCGGGACGCCATCAAGATGCTGGACGACGCCTCCACGTCTGCGAAGAAGCTGCGGAACAAGGCGAACTATCTGTCGCACGAGCTCGAGCTGTTCGAAGAGGCCTTCCTCAAGCACAATAATTAA
Protein
MAAYVNRTISMMAGDGPHNVSMLNNGSVRVNMQNVDSPLQIFVRAKKKINDIFVEIDDYVKDAVTFMHAVSGENGIATPQDMGNVESYVSKVEAIREVLKRDHMKVAFFGRTSNGKSTVINAMLHDKILPSGIGHTTNCFLQVEGSDTNEAYMRTEGCEEKLNVQSVSQLGHALCATRLQECSLVHVHWPRELCALLRDDVVLVDSPGVDVTPNLDTWIDKYCLDADVFVLVANAESTLMVTEKNFFHKVSTKISQPNIFILNNRWDASASEPEYMEQVRTQHANRCVDFLSRELRVCSPKEAEERIFFISAKEALLTRMRDREKPVSSPILAEGHQVRYFEFVDFERKFEECISQSAVRTKFAQHSRRGKNIAGDVMAALDRVYNIATEQKAAKVEKQRILHEQLSSIEEQLTTITRQMKDKINRMVESVEHKVSLTLSQEIRRLSALVDEYESEFRPERPALEQYKRALHRHVEAGLGSRLKKRLSSDIGNEMDVVQKEMAERMYNILPTNKRAAAANYIIPHQQPFEVLYRLNCDNLCADFNEDLSFRFSYGITALIQRFQGKNTNRIALNNPPQYTQIPASITPVSALPPAPRAALPVLSAEEWGLLSKFALGALTSQGTMGGLLLSGLLLKTVGWRVLAVTGLAYGALYAYERLTWTAKAQERVFKRQYVAHAGKKLRLIVDLTSANCSHQVQQELSTMFARLCRLIDEATTEMDAELSGVRDAIKMLDDASTSAKKLRNKANYLSHELELFEEAFLKHNN
Summary
Description
Essential transmembrane GTPase, which mediates mitochondrial fusion. Fusion of mitochondria occurs in many cell types and constitutes an important step in mitochondria morphology, which is balanced between fusion and fission (By similarity).
Catalytic Activity
GTP + H2O = GDP + H(+) + phosphate
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family. Mitofusin subfamily.
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family. Mitofusin subfamily.
Keywords
Alternative splicing
Coiled coil
Complete proteome
GTP-binding
Hydrolase
Membrane
Mitochondrion
Mitochondrion outer membrane
Nucleotide-binding
Phosphoprotein
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Transmembrane GTPase Marf
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9IZY5
A0A194Q3Q4
A0A212EHK1
A0A2A4JN98
A0A0N0U5N6
A0A0L7QU79
+ More
A0A088A1V1 A0A232EUT0 D6X0Y4 K7J2Q7 A0A1B6EQB9 A0A1Y1M5B5 A0A151XDH4 A0A0C9QF62 E2AJ70 A0A195C0R9 A0A154PSK0 A0A2A3ELQ1 E2C5A5 A0A2J7RLY1 A0A067RG51 A0A2J7Q823 A0A151JZ80 A0A2J7Q826 U4U0M6 E0W1N1 U4U7P1 A0A069DW95 A0A224XKA2 A0A1B6FXW6 A0A1B6E5A2 A0A0V0G5D8 A0A1B6DM15 A0A026WUY2 T1HN33 A0A3L8D8E0 A0A0P5KIR2 A0A0N8CFU0 A0A182XV89 E9GGH3 A0A336LLH5 U5EVN5 A0A0K8TM46 A0A182JPZ7 A0A1B6KHF7 A0A084VAJ8 A0A182VRI9 A0A182MLY6 W8BLS3 A0A0K8VW17 A0A182Q5C8 W5JL04 A0A2M4A406 A0A182HGT0 A0A336K1Q3 A0A182NFK2 A0A182VEB8 A0A182U957 A0A182P9S6 Q7PY95 A0A182F6U5 A0A182X2R1 A0A1B6L1W0 T1JMD1 A0A2M4BEN0 A0A2M3Z1T2 A0A0P6CWD6 A0A0M3QZD3 A0A2M4BEW3 A0A1B6DDV5 A0A034VE49 A0A182RNZ3 A0A0P6EBP7 A0A182JIE9 A0A1A9WT32 T1P9C3 Q7YU24 B4L2G6 A0A1A9XYI5 A0A1B0ASS6 A0A1A9VWY0 A0A1A9Z3W2 B4PYS3 B3NXS3 A0A1A9YUD3 F4X1N2 Q7YU24-2 A0A0R1E9R8 A0A182H8J7 A0A1S4F7U9 A0A0Q5T5B2 B4M7X8 A0A023EVU9 A0A0Q9WZD8 Q29IJ1 B3MXY1 Q17CQ1 A0A1Q3FFY5 B4JNY8
A0A088A1V1 A0A232EUT0 D6X0Y4 K7J2Q7 A0A1B6EQB9 A0A1Y1M5B5 A0A151XDH4 A0A0C9QF62 E2AJ70 A0A195C0R9 A0A154PSK0 A0A2A3ELQ1 E2C5A5 A0A2J7RLY1 A0A067RG51 A0A2J7Q823 A0A151JZ80 A0A2J7Q826 U4U0M6 E0W1N1 U4U7P1 A0A069DW95 A0A224XKA2 A0A1B6FXW6 A0A1B6E5A2 A0A0V0G5D8 A0A1B6DM15 A0A026WUY2 T1HN33 A0A3L8D8E0 A0A0P5KIR2 A0A0N8CFU0 A0A182XV89 E9GGH3 A0A336LLH5 U5EVN5 A0A0K8TM46 A0A182JPZ7 A0A1B6KHF7 A0A084VAJ8 A0A182VRI9 A0A182MLY6 W8BLS3 A0A0K8VW17 A0A182Q5C8 W5JL04 A0A2M4A406 A0A182HGT0 A0A336K1Q3 A0A182NFK2 A0A182VEB8 A0A182U957 A0A182P9S6 Q7PY95 A0A182F6U5 A0A182X2R1 A0A1B6L1W0 T1JMD1 A0A2M4BEN0 A0A2M3Z1T2 A0A0P6CWD6 A0A0M3QZD3 A0A2M4BEW3 A0A1B6DDV5 A0A034VE49 A0A182RNZ3 A0A0P6EBP7 A0A182JIE9 A0A1A9WT32 T1P9C3 Q7YU24 B4L2G6 A0A1A9XYI5 A0A1B0ASS6 A0A1A9VWY0 A0A1A9Z3W2 B4PYS3 B3NXS3 A0A1A9YUD3 F4X1N2 Q7YU24-2 A0A0R1E9R8 A0A182H8J7 A0A1S4F7U9 A0A0Q5T5B2 B4M7X8 A0A023EVU9 A0A0Q9WZD8 Q29IJ1 B3MXY1 Q17CQ1 A0A1Q3FFY5 B4JNY8
EC Number
3.6.5.-
Pubmed
19121390
26354079
22118469
28648823
18362917
19820115
+ More
20075255 28004739 20798317 24845553 23537049 20566863 26334808 24508170 30249741 25244985 21292972 26369729 24438588 24495485 20920257 23761445 12364791 14747013 17210077 25348373 25315136 12598526 10731132 12537572 12537569 12128227 18327897 17994087 17550304 18057021 21719571 26483478 24945155 15632085 17510324
20075255 28004739 20798317 24845553 23537049 20566863 26334808 24508170 30249741 25244985 21292972 26369729 24438588 24495485 20920257 23761445 12364791 14747013 17210077 25348373 25315136 12598526 10731132 12537572 12537569 12128227 18327897 17994087 17550304 18057021 21719571 26483478 24945155 15632085 17510324
EMBL
BABH01018950
KQ459472
KPJ00168.1
AGBW02014864
OWR40973.1
NWSH01001058
+ More
PCG72880.1 KQ435776 KOX74882.1 KQ414735 KOC62175.1 NNAY01002089 OXU22112.1 KQ971372 EFA09534.1 GECZ01029624 GECZ01006188 GECZ01002613 JAS40145.1 JAS63581.1 JAS67156.1 GEZM01041604 GEZM01041603 JAV80258.1 KQ982294 KYQ58403.1 GBYB01013141 GBYB01013142 GBYB01013143 JAG82908.1 JAG82909.1 JAG82910.1 GL439967 EFN66510.1 KQ978379 KYM94484.1 KQ435050 KZC14238.1 KZ288215 PBC32685.1 GL452770 EFN76838.1 NEVH01002587 PNF41840.1 KK852705 KDR18029.1 NEVH01016978 PNF24730.1 KQ981432 KYN41680.1 PNF24729.1 KB631843 ERL86642.1 DS235872 EEB19537.1 ERL86641.1 GBGD01000561 JAC88328.1 GFTR01007987 JAW08439.1 GECZ01014731 JAS55038.1 GEDC01004221 GEDC01002891 JAS33077.1 JAS34407.1 GECL01002826 JAP03298.1 GEDC01010579 JAS26719.1 KK107111 EZA58914.1 ACPB03015200 QOIP01000011 RLU16551.1 GDIQ01209141 JAK42584.1 GDIP01135927 LRGB01000725 JAL67787.1 KZS16364.1 GL732543 EFX81498.1 UFQS01000045 UFQT01000045 SSW98420.1 SSX18806.1 GANO01000940 JAB58931.1 GDAI01002402 JAI15201.1 GEBQ01029641 GEBQ01029091 JAT10336.1 JAT10886.1 ATLV01004127 KE524190 KFB34992.1 AXCM01002852 GAMC01004530 GAMC01004529 JAC02026.1 GDHF01009240 JAI43074.1 AXCN02000208 ADMH02001255 ETN63434.1 GGFK01002139 MBW35460.1 APCN01005945 SSW98421.1 SSX18807.1 AAAB01008987 EAA01124.5 EGK97464.1 GEBQ01022356 GEBQ01016652 JAT17621.1 JAT23325.1 JH431842 GGFJ01002351 MBW51492.1 GGFM01001741 MBW22492.1 GDIQ01087584 JAN07153.1 CP012528 ALC49223.1 GGFJ01002350 MBW51491.1 GEDC01013523 JAS23775.1 GAKP01018566 GAKP01018565 JAC40387.1 GDIQ01070838 JAN23899.1 KA645326 KA645327 AFP59956.1 AF355475 AE014298 AY095019 BT010027 CH933810 EDW06842.1 JXJN01002979 CM000162 EDX01009.1 KRK05878.1 CH954180 EDV47374.1 KQS30466.1 CCAG010019883 GL888538 EGI59639.1 KRK05877.1 JXUM01118571 JXUM01118572 KQ566201 KXJ70328.1 KQS30467.1 CH940653 EDW62895.1 GAPW01000255 JAC13343.1 KRF93834.1 CH379063 EAL32662.2 CH902630 EDV38596.1 CH477305 EAT44145.1 GFDL01008577 JAV26468.1 CH916371 EDV92431.1
PCG72880.1 KQ435776 KOX74882.1 KQ414735 KOC62175.1 NNAY01002089 OXU22112.1 KQ971372 EFA09534.1 GECZ01029624 GECZ01006188 GECZ01002613 JAS40145.1 JAS63581.1 JAS67156.1 GEZM01041604 GEZM01041603 JAV80258.1 KQ982294 KYQ58403.1 GBYB01013141 GBYB01013142 GBYB01013143 JAG82908.1 JAG82909.1 JAG82910.1 GL439967 EFN66510.1 KQ978379 KYM94484.1 KQ435050 KZC14238.1 KZ288215 PBC32685.1 GL452770 EFN76838.1 NEVH01002587 PNF41840.1 KK852705 KDR18029.1 NEVH01016978 PNF24730.1 KQ981432 KYN41680.1 PNF24729.1 KB631843 ERL86642.1 DS235872 EEB19537.1 ERL86641.1 GBGD01000561 JAC88328.1 GFTR01007987 JAW08439.1 GECZ01014731 JAS55038.1 GEDC01004221 GEDC01002891 JAS33077.1 JAS34407.1 GECL01002826 JAP03298.1 GEDC01010579 JAS26719.1 KK107111 EZA58914.1 ACPB03015200 QOIP01000011 RLU16551.1 GDIQ01209141 JAK42584.1 GDIP01135927 LRGB01000725 JAL67787.1 KZS16364.1 GL732543 EFX81498.1 UFQS01000045 UFQT01000045 SSW98420.1 SSX18806.1 GANO01000940 JAB58931.1 GDAI01002402 JAI15201.1 GEBQ01029641 GEBQ01029091 JAT10336.1 JAT10886.1 ATLV01004127 KE524190 KFB34992.1 AXCM01002852 GAMC01004530 GAMC01004529 JAC02026.1 GDHF01009240 JAI43074.1 AXCN02000208 ADMH02001255 ETN63434.1 GGFK01002139 MBW35460.1 APCN01005945 SSW98421.1 SSX18807.1 AAAB01008987 EAA01124.5 EGK97464.1 GEBQ01022356 GEBQ01016652 JAT17621.1 JAT23325.1 JH431842 GGFJ01002351 MBW51492.1 GGFM01001741 MBW22492.1 GDIQ01087584 JAN07153.1 CP012528 ALC49223.1 GGFJ01002350 MBW51491.1 GEDC01013523 JAS23775.1 GAKP01018566 GAKP01018565 JAC40387.1 GDIQ01070838 JAN23899.1 KA645326 KA645327 AFP59956.1 AF355475 AE014298 AY095019 BT010027 CH933810 EDW06842.1 JXJN01002979 CM000162 EDX01009.1 KRK05878.1 CH954180 EDV47374.1 KQS30466.1 CCAG010019883 GL888538 EGI59639.1 KRK05877.1 JXUM01118571 JXUM01118572 KQ566201 KXJ70328.1 KQS30467.1 CH940653 EDW62895.1 GAPW01000255 JAC13343.1 KRF93834.1 CH379063 EAL32662.2 CH902630 EDV38596.1 CH477305 EAT44145.1 GFDL01008577 JAV26468.1 CH916371 EDV92431.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000053105
UP000053825
+ More
UP000005203 UP000215335 UP000007266 UP000002358 UP000075809 UP000000311 UP000078542 UP000076502 UP000242457 UP000008237 UP000235965 UP000027135 UP000078541 UP000030742 UP000009046 UP000053097 UP000015103 UP000279307 UP000076858 UP000076408 UP000000305 UP000075881 UP000030765 UP000075920 UP000075883 UP000075886 UP000000673 UP000075840 UP000075884 UP000075903 UP000075902 UP000075885 UP000007062 UP000069272 UP000076407 UP000092553 UP000075900 UP000075880 UP000091820 UP000095301 UP000000803 UP000009192 UP000092443 UP000092460 UP000078200 UP000092445 UP000002282 UP000008711 UP000092444 UP000007755 UP000069940 UP000249989 UP000008792 UP000001819 UP000007801 UP000008820 UP000001070
UP000005203 UP000215335 UP000007266 UP000002358 UP000075809 UP000000311 UP000078542 UP000076502 UP000242457 UP000008237 UP000235965 UP000027135 UP000078541 UP000030742 UP000009046 UP000053097 UP000015103 UP000279307 UP000076858 UP000076408 UP000000305 UP000075881 UP000030765 UP000075920 UP000075883 UP000075886 UP000000673 UP000075840 UP000075884 UP000075903 UP000075902 UP000075885 UP000007062 UP000069272 UP000076407 UP000092553 UP000075900 UP000075880 UP000091820 UP000095301 UP000000803 UP000009192 UP000092443 UP000092460 UP000078200 UP000092445 UP000002282 UP000008711 UP000092444 UP000007755 UP000069940 UP000249989 UP000008792 UP000001819 UP000007801 UP000008820 UP000001070
Interpro
ProteinModelPortal
H9IZY5
A0A194Q3Q4
A0A212EHK1
A0A2A4JN98
A0A0N0U5N6
A0A0L7QU79
+ More
A0A088A1V1 A0A232EUT0 D6X0Y4 K7J2Q7 A0A1B6EQB9 A0A1Y1M5B5 A0A151XDH4 A0A0C9QF62 E2AJ70 A0A195C0R9 A0A154PSK0 A0A2A3ELQ1 E2C5A5 A0A2J7RLY1 A0A067RG51 A0A2J7Q823 A0A151JZ80 A0A2J7Q826 U4U0M6 E0W1N1 U4U7P1 A0A069DW95 A0A224XKA2 A0A1B6FXW6 A0A1B6E5A2 A0A0V0G5D8 A0A1B6DM15 A0A026WUY2 T1HN33 A0A3L8D8E0 A0A0P5KIR2 A0A0N8CFU0 A0A182XV89 E9GGH3 A0A336LLH5 U5EVN5 A0A0K8TM46 A0A182JPZ7 A0A1B6KHF7 A0A084VAJ8 A0A182VRI9 A0A182MLY6 W8BLS3 A0A0K8VW17 A0A182Q5C8 W5JL04 A0A2M4A406 A0A182HGT0 A0A336K1Q3 A0A182NFK2 A0A182VEB8 A0A182U957 A0A182P9S6 Q7PY95 A0A182F6U5 A0A182X2R1 A0A1B6L1W0 T1JMD1 A0A2M4BEN0 A0A2M3Z1T2 A0A0P6CWD6 A0A0M3QZD3 A0A2M4BEW3 A0A1B6DDV5 A0A034VE49 A0A182RNZ3 A0A0P6EBP7 A0A182JIE9 A0A1A9WT32 T1P9C3 Q7YU24 B4L2G6 A0A1A9XYI5 A0A1B0ASS6 A0A1A9VWY0 A0A1A9Z3W2 B4PYS3 B3NXS3 A0A1A9YUD3 F4X1N2 Q7YU24-2 A0A0R1E9R8 A0A182H8J7 A0A1S4F7U9 A0A0Q5T5B2 B4M7X8 A0A023EVU9 A0A0Q9WZD8 Q29IJ1 B3MXY1 Q17CQ1 A0A1Q3FFY5 B4JNY8
A0A088A1V1 A0A232EUT0 D6X0Y4 K7J2Q7 A0A1B6EQB9 A0A1Y1M5B5 A0A151XDH4 A0A0C9QF62 E2AJ70 A0A195C0R9 A0A154PSK0 A0A2A3ELQ1 E2C5A5 A0A2J7RLY1 A0A067RG51 A0A2J7Q823 A0A151JZ80 A0A2J7Q826 U4U0M6 E0W1N1 U4U7P1 A0A069DW95 A0A224XKA2 A0A1B6FXW6 A0A1B6E5A2 A0A0V0G5D8 A0A1B6DM15 A0A026WUY2 T1HN33 A0A3L8D8E0 A0A0P5KIR2 A0A0N8CFU0 A0A182XV89 E9GGH3 A0A336LLH5 U5EVN5 A0A0K8TM46 A0A182JPZ7 A0A1B6KHF7 A0A084VAJ8 A0A182VRI9 A0A182MLY6 W8BLS3 A0A0K8VW17 A0A182Q5C8 W5JL04 A0A2M4A406 A0A182HGT0 A0A336K1Q3 A0A182NFK2 A0A182VEB8 A0A182U957 A0A182P9S6 Q7PY95 A0A182F6U5 A0A182X2R1 A0A1B6L1W0 T1JMD1 A0A2M4BEN0 A0A2M3Z1T2 A0A0P6CWD6 A0A0M3QZD3 A0A2M4BEW3 A0A1B6DDV5 A0A034VE49 A0A182RNZ3 A0A0P6EBP7 A0A182JIE9 A0A1A9WT32 T1P9C3 Q7YU24 B4L2G6 A0A1A9XYI5 A0A1B0ASS6 A0A1A9VWY0 A0A1A9Z3W2 B4PYS3 B3NXS3 A0A1A9YUD3 F4X1N2 Q7YU24-2 A0A0R1E9R8 A0A182H8J7 A0A1S4F7U9 A0A0Q5T5B2 B4M7X8 A0A023EVU9 A0A0Q9WZD8 Q29IJ1 B3MXY1 Q17CQ1 A0A1Q3FFY5 B4JNY8
PDB
5GOM
E-value=1.93319e-110,
Score=1023
Ontologies
PATHWAY
GO
PANTHER
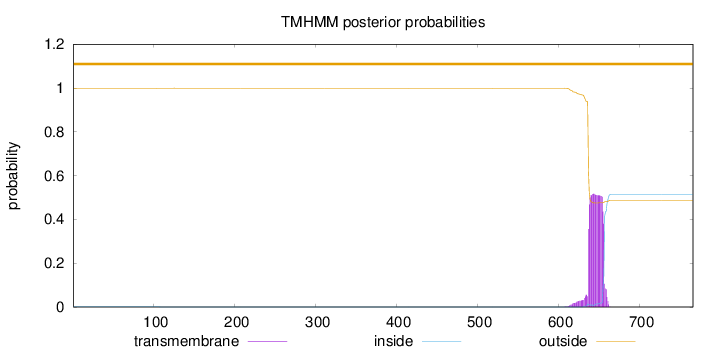
Topology
Subcellular location
Mitochondrion outer membrane
Length:
766
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.81101
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.00066
outside
1 - 766
Population Genetic Test Statistics
Pi
16.913195
Theta
20.545314
Tajima's D
-0.512399
CLR
1.120078
CSRT
0.24208789560522
Interpretation
Uncertain
