Gene
KWMTBOMO05997
Pre Gene Modal
BGIBMGA002819
Annotation
PREDICTED:_WD_repeat-containing_protein_66-like_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.405 PlasmaMembrane Reliability : 1.636
Sequence
CDS
ATGGCGACTATTTTTTCAGGCAACACACCTTGCCCGTGTTGGGAAAAAGTTGTAGCTGAGATCGATTCAAACACAGGATTATTAAAACAGAAGTTGATAAAGAAGAAACTGCCAAGTGACGCTACTGTCAGCGGGAAGACTTTCCTTGTCAGGGATTTTATTGTTTGTACAAACAACCAAGGTGTCGGTTTCGTTGATTTCGCAACTGAGAAGCTTGTCACAGTTTTGAACAGCAAACGGTCGTATGCTTTAGCATTGAGTGTGCATCCAGAGAAGCCATATGTGTGCATCGGATACGACAATGGTATAGTTGAAATGTATAATTTTGTTCAGCACAAGCTGTTCGTTAGACTCGACCTGAGAGATTTGTACAAGATCGTCATCCCTCCAAAAGACGACTCCATAAAATGCAACTACGAAGTAACTGTACCTCCGATTTCTGTGTCGTGTTTGAAATATTCTCATTCAGGAATGCATTTAGCTTGCGGACTGAACACAGGACAGTTGCTGTTCTTACATCCAACGACAATAGACGTTATAACCGAAATTCCATACACGGACACCGGGCACGCCATCAAACAGATTACGTATAGCTGCGATTCACTAACGCTCGCTCTGTTTGATGCTGGTCGAACAGTATGCGTTTACAAATACGACTGCCAGGATTTATCATGGAAATTCATTGGCAAACATCGAGCTCACTACAAAGATATTACATCAATATTATTCCTCTCAGAAAAGAATGCAAACGGAGAATATAAGCTTATATCTTTGGGGATCGATAGAATTATGGTAGAATATGATATCGGAGAGAGTGCCGACGGCTATTTGGAAATACTCAGCTTGGACAGACTGGATCAGACCGCTGTACCATTAGCCGGGATTCAGTGGCCGTCACCTGAAGGCCTGGATCCGGATGTGCACAGATTAGATCTGCCTTTGATTCTGGTTGCTAATGATGAGTTCAAATACAAAATAGTGAACTACGCAACTACGATGACTTTATCGACAATATTGGGCCCTCGATATGAGCATCCAGTACGCAGAATGTTATTGACTGCAAGAGAGGAAGATAACGGAAAGCAACAATACTTGATATTTGCCACCAAGAACGTTGTGGGGATACAGAAGATGCCCATAGATGGTAATCCTTGGAAGCACGCGGGCTTGCTTGGACATCCATTGGAGGTAACAGAGATGTGTTTCAGGGAAGACTGTGGCATACTGTTTACGATTGGAGACAAAGATGGATGCATGAGTCAATGGGCTATTAATTACAGATCTGTAGACACCACGACGAAACAAGGGGGCGGGGACTTAGATCCGTATTACTGTCTGATCGAGAACGGGCGTCCGGGCTGGCTCTTCCAAGAAATTCGTGATTTATTCTACTACATACAGATATTGTGTCAGGGCACGTTCTCGCCCGCAATGAGGAGAGTCAAAGATTTTATACCCATTGATTCGCTACCGGATCTAATGAGAGCGCTGGGATTTTTCTTATCTGAATATGAGACAGAAAATTTAATTGCAGAAGCGAAATACAAGGTGTATCAAATTAATCCATCGACCGAAATAGACTTTGAGGAGTTTGTGAAGCTGTACATCAATCATCGGCCTGCATTCGGGTACAGATACAAGAAAATACGTGACGCTTTTGCACACTTCGCGTATTTGGATAATGAACAGTTGGCCATAAACAGGAACGGGTTGATTGAGCTGCTTGCGGATTATGGTGAACGTTTCCCTCGGGAATTGACTTGGTATTTAATATCAATATTGTGTGGGGAGAGCTTCGAAGACAGAGCGAAGATGGCTGAAAACGATTTTTCGTTTATGCCAGAGGAGCTGACTCTTGACTATTTAGCCACCTACGTCATAGGTGTCGATGACTTGGAAAACATTCCCGAGGGATCTTCAGGGTTGGGGTCGTTCGACACACAATCGGCACAGACGAGTACTACGGGAGACTGA
Protein
MATIFSGNTPCPCWEKVVAEIDSNTGLLKQKLIKKKLPSDATVSGKTFLVRDFIVCTNNQGVGFVDFATEKLVTVLNSKRSYALALSVHPEKPYVCIGYDNGIVEMYNFVQHKLFVRLDLRDLYKIVIPPKDDSIKCNYEVTVPPISVSCLKYSHSGMHLACGLNTGQLLFLHPTTIDVITEIPYTDTGHAIKQITYSCDSLTLALFDAGRTVCVYKYDCQDLSWKFIGKHRAHYKDITSILFLSEKNANGEYKLISLGIDRIMVEYDIGESADGYLEILSLDRLDQTAVPLAGIQWPSPEGLDPDVHRLDLPLILVANDEFKYKIVNYATTMTLSTILGPRYEHPVRRMLLTAREEDNGKQQYLIFATKNVVGIQKMPIDGNPWKHAGLLGHPLEVTEMCFREDCGILFTIGDKDGCMSQWAINYRSVDTTTKQGGGDLDPYYCLIENGRPGWLFQEIRDLFYYIQILCQGTFSPAMRRVKDFIPIDSLPDLMRALGFFLSEYETENLIAEAKYKVYQINPSTEIDFEEFVKLYINHRPAFGYRYKKIRDAFAHFAYLDNEQLAINRNGLIELLADYGERFPRELTWYLISILCGESFEDRAKMAENDFSFMPEELTLDYLATYVIGVDDLENIPEGSSGLGSFDTQSAQTSTTGD
Summary
Uniprot
A0A194QR62
A0A212EHL8
A0A3S2TTN8
A0A194Q9S4
A0A2H1VV69
A0A2A4JN92
+ More
A0A139WCL9 K7IXD4 A0A154P067 A0A151XI81 A0A310SPW8 A0A232EV42 W5MD80 H2YNA2 A0A026WAQ2 A0A2A3EHE8 A0A1L8I155 A0A195EQ42 A0A087ZPP4 F7ABS5 A0A3Q1IBX8 A0A3Q0K5M8 V4BRV3 A0A151I7P7 A0A210QD55 F7EE82 A0A3P8W6T1 K1Q7E1 A0A1I8HI71 W5MDA3 A0A267GMP3 R7UDH9 A0A1I8JEL0 A0A060XJ43 A0A267GPP3 B3RYZ2 F1Q8Y1 A0A267DJZ0 A0A2R8RY46 A0A369S1V2 G3VUV1 A0A3B4CV03 A0A3B4ZRR4 C3XUL0 A0A3Q1C600 K7G9M2 A3QK38 A0A151NSZ7 K7G9P0 A0A091NQT4 A0A1U7RLX3 A0A3Q1F350 A0A3B4VE41 F7FU35 A0A3B4Y5N2 A0A093ISJ5 A0A087VDC4 A0A1S3K2G4 A0A2D0PRQ2 A0A1S3K1S6 A0A1I8GK17 A0A3P8UDA3 A0A1W4ZXE4 A0A1X7VIL7 A0A091U632 A0A094KT60 A0A091PNI1 A0A091KE96 M4ASZ2 A0A3Q3BAU9 A0A091SNI9 A0A091HKZ6 F6VXV4 A0A2I0M896 H0Z6W3 A0A3Q3XA48 A0A093Q8R4 A0A0A0A5U5 A0A099ZT44 A0A3S2P745 A0A3L8SD17 A0A1V4JS62 A0A146Y6I4 A0A3Q2NQ52 A0A087RB13 A0A2I4BYH7 A0A147AS80 A0A3P8YJC0 A0A3M0K3H5 A0A093HE96 A0A218V5J8 A0A1A8B005 A0A3Q2Z6U6 A0A091JLX1 H3BG34 A0A1V4JS94 A0A091GXZ8 A0A093NM27 A0A091WM90
A0A139WCL9 K7IXD4 A0A154P067 A0A151XI81 A0A310SPW8 A0A232EV42 W5MD80 H2YNA2 A0A026WAQ2 A0A2A3EHE8 A0A1L8I155 A0A195EQ42 A0A087ZPP4 F7ABS5 A0A3Q1IBX8 A0A3Q0K5M8 V4BRV3 A0A151I7P7 A0A210QD55 F7EE82 A0A3P8W6T1 K1Q7E1 A0A1I8HI71 W5MDA3 A0A267GMP3 R7UDH9 A0A1I8JEL0 A0A060XJ43 A0A267GPP3 B3RYZ2 F1Q8Y1 A0A267DJZ0 A0A2R8RY46 A0A369S1V2 G3VUV1 A0A3B4CV03 A0A3B4ZRR4 C3XUL0 A0A3Q1C600 K7G9M2 A3QK38 A0A151NSZ7 K7G9P0 A0A091NQT4 A0A1U7RLX3 A0A3Q1F350 A0A3B4VE41 F7FU35 A0A3B4Y5N2 A0A093ISJ5 A0A087VDC4 A0A1S3K2G4 A0A2D0PRQ2 A0A1S3K1S6 A0A1I8GK17 A0A3P8UDA3 A0A1W4ZXE4 A0A1X7VIL7 A0A091U632 A0A094KT60 A0A091PNI1 A0A091KE96 M4ASZ2 A0A3Q3BAU9 A0A091SNI9 A0A091HKZ6 F6VXV4 A0A2I0M896 H0Z6W3 A0A3Q3XA48 A0A093Q8R4 A0A0A0A5U5 A0A099ZT44 A0A3S2P745 A0A3L8SD17 A0A1V4JS62 A0A146Y6I4 A0A3Q2NQ52 A0A087RB13 A0A2I4BYH7 A0A147AS80 A0A3P8YJC0 A0A3M0K3H5 A0A093HE96 A0A218V5J8 A0A1A8B005 A0A3Q2Z6U6 A0A091JLX1 H3BG34 A0A1V4JS94 A0A091GXZ8 A0A093NM27 A0A091WM90
Pubmed
EMBL
KQ461175
KPJ07993.1
AGBW02014864
OWR40975.1
RSAL01000002
RVE54958.1
+ More
KQ459472 KPJ00166.1 ODYU01004342 SOQ44114.1 NWSH01001058 PCG72882.1 KQ971365 KYB25656.1 AAZX01006967 KQ434782 KZC04530.1 KQ982082 KYQ60114.1 KQ761824 OAD56770.1 NNAY01002061 OXU22203.1 AHAT01005122 KK107293 EZA53167.1 KZ288254 PBC30712.1 CM004466 OCU02055.1 KQ982021 KYN30405.1 KB202237 ESO91649.1 KQ978397 KYM94219.1 NEDP02004104 OWF46674.1 AAMC01072987 AAMC01072988 AAMC01072989 JH816920 EKC24770.1 NIVC01000238 PAA87286.1 AMQN01001471 AMQN01001472 KB302615 ELU04166.1 FR905184 CDQ76910.1 NIVC01000208 PAA87973.1 DS985246 EDV24111.1 CR931763 NIVC01004035 PAA48859.1 CR848794 NOWV01000093 RDD40709.1 AEFK01071537 AEFK01071538 AEFK01071539 AEFK01071540 AEFK01071541 GG666465 EEN68292.1 AGCU01088573 AGCU01088574 AGCU01088575 AGCU01088576 AGCU01088577 AKHW03002185 KYO39729.1 KL391157 KFP91671.1 KK600277 KFW05695.1 KL489047 KFO10616.1 KK419334 KFQ86164.1 KL356336 KFZ62428.1 KK663160 KFQ09467.1 KK741510 KFP38904.1 KK478981 KFQ59903.1 KL217497 KFO95787.1 AKCR02000030 PKK25901.1 ABQF01047133 KL415735 KFW80562.1 KL870823 KGL89416.1 KL897916 KGL84942.1 CM012445 RVE68237.1 QUSF01000030 RLV99937.1 LSYS01006629 OPJ75032.1 GCES01036707 JAR49616.1 KL226265 KFM10667.1 GCES01004890 JAR81433.1 QRBI01000121 RMC05774.1 KL217402 KFV77307.1 MUZQ01000045 OWK61246.1 HADY01022287 SBP60772.1 KK501101 KFP12701.1 AFYH01016466 AFYH01016467 AFYH01016468 AFYH01016469 AFYH01016470 OPJ75031.1 KL513722 KFO87310.1 KL224874 KFW65156.1 KK735845 KFR16235.1
KQ459472 KPJ00166.1 ODYU01004342 SOQ44114.1 NWSH01001058 PCG72882.1 KQ971365 KYB25656.1 AAZX01006967 KQ434782 KZC04530.1 KQ982082 KYQ60114.1 KQ761824 OAD56770.1 NNAY01002061 OXU22203.1 AHAT01005122 KK107293 EZA53167.1 KZ288254 PBC30712.1 CM004466 OCU02055.1 KQ982021 KYN30405.1 KB202237 ESO91649.1 KQ978397 KYM94219.1 NEDP02004104 OWF46674.1 AAMC01072987 AAMC01072988 AAMC01072989 JH816920 EKC24770.1 NIVC01000238 PAA87286.1 AMQN01001471 AMQN01001472 KB302615 ELU04166.1 FR905184 CDQ76910.1 NIVC01000208 PAA87973.1 DS985246 EDV24111.1 CR931763 NIVC01004035 PAA48859.1 CR848794 NOWV01000093 RDD40709.1 AEFK01071537 AEFK01071538 AEFK01071539 AEFK01071540 AEFK01071541 GG666465 EEN68292.1 AGCU01088573 AGCU01088574 AGCU01088575 AGCU01088576 AGCU01088577 AKHW03002185 KYO39729.1 KL391157 KFP91671.1 KK600277 KFW05695.1 KL489047 KFO10616.1 KK419334 KFQ86164.1 KL356336 KFZ62428.1 KK663160 KFQ09467.1 KK741510 KFP38904.1 KK478981 KFQ59903.1 KL217497 KFO95787.1 AKCR02000030 PKK25901.1 ABQF01047133 KL415735 KFW80562.1 KL870823 KGL89416.1 KL897916 KGL84942.1 CM012445 RVE68237.1 QUSF01000030 RLV99937.1 LSYS01006629 OPJ75032.1 GCES01036707 JAR49616.1 KL226265 KFM10667.1 GCES01004890 JAR81433.1 QRBI01000121 RMC05774.1 KL217402 KFV77307.1 MUZQ01000045 OWK61246.1 HADY01022287 SBP60772.1 KK501101 KFP12701.1 AFYH01016466 AFYH01016467 AFYH01016468 AFYH01016469 AFYH01016470 OPJ75031.1 KL513722 KFO87310.1 KL224874 KFW65156.1 KK735845 KFR16235.1
Proteomes
UP000053240
UP000007151
UP000283053
UP000053268
UP000218220
UP000007266
+ More
UP000002358 UP000076502 UP000075809 UP000215335 UP000018468 UP000007875 UP000053097 UP000242457 UP000186698 UP000078541 UP000005203 UP000008144 UP000265040 UP000030746 UP000078542 UP000242188 UP000008143 UP000265120 UP000005408 UP000095280 UP000215902 UP000014760 UP000193380 UP000009022 UP000000437 UP000253843 UP000007648 UP000261440 UP000261400 UP000001554 UP000257160 UP000007267 UP000050525 UP000189705 UP000257200 UP000261420 UP000002280 UP000261360 UP000085678 UP000221080 UP000265080 UP000192224 UP000007879 UP000002852 UP000264800 UP000054308 UP000002279 UP000053872 UP000007754 UP000261620 UP000053858 UP000053641 UP000276834 UP000190648 UP000265000 UP000053286 UP000192220 UP000265140 UP000269221 UP000053875 UP000197619 UP000264820 UP000053119 UP000008672 UP000054081 UP000053605
UP000002358 UP000076502 UP000075809 UP000215335 UP000018468 UP000007875 UP000053097 UP000242457 UP000186698 UP000078541 UP000005203 UP000008144 UP000265040 UP000030746 UP000078542 UP000242188 UP000008143 UP000265120 UP000005408 UP000095280 UP000215902 UP000014760 UP000193380 UP000009022 UP000000437 UP000253843 UP000007648 UP000261440 UP000261400 UP000001554 UP000257160 UP000007267 UP000050525 UP000189705 UP000257200 UP000261420 UP000002280 UP000261360 UP000085678 UP000221080 UP000265080 UP000192224 UP000007879 UP000002852 UP000264800 UP000054308 UP000002279 UP000053872 UP000007754 UP000261620 UP000053858 UP000053641 UP000276834 UP000190648 UP000265000 UP000053286 UP000192220 UP000265140 UP000269221 UP000053875 UP000197619 UP000264820 UP000053119 UP000008672 UP000054081 UP000053605
Interpro
IPR011992
EF-hand-dom_pair
+ More
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR036034 PDZ_sf
IPR001478 PDZ
IPR040815 Nas2_N
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR019775 WD40_repeat_CS
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR036034 PDZ_sf
IPR001478 PDZ
IPR040815 Nas2_N
SUPFAM
Gene 3D
ProteinModelPortal
A0A194QR62
A0A212EHL8
A0A3S2TTN8
A0A194Q9S4
A0A2H1VV69
A0A2A4JN92
+ More
A0A139WCL9 K7IXD4 A0A154P067 A0A151XI81 A0A310SPW8 A0A232EV42 W5MD80 H2YNA2 A0A026WAQ2 A0A2A3EHE8 A0A1L8I155 A0A195EQ42 A0A087ZPP4 F7ABS5 A0A3Q1IBX8 A0A3Q0K5M8 V4BRV3 A0A151I7P7 A0A210QD55 F7EE82 A0A3P8W6T1 K1Q7E1 A0A1I8HI71 W5MDA3 A0A267GMP3 R7UDH9 A0A1I8JEL0 A0A060XJ43 A0A267GPP3 B3RYZ2 F1Q8Y1 A0A267DJZ0 A0A2R8RY46 A0A369S1V2 G3VUV1 A0A3B4CV03 A0A3B4ZRR4 C3XUL0 A0A3Q1C600 K7G9M2 A3QK38 A0A151NSZ7 K7G9P0 A0A091NQT4 A0A1U7RLX3 A0A3Q1F350 A0A3B4VE41 F7FU35 A0A3B4Y5N2 A0A093ISJ5 A0A087VDC4 A0A1S3K2G4 A0A2D0PRQ2 A0A1S3K1S6 A0A1I8GK17 A0A3P8UDA3 A0A1W4ZXE4 A0A1X7VIL7 A0A091U632 A0A094KT60 A0A091PNI1 A0A091KE96 M4ASZ2 A0A3Q3BAU9 A0A091SNI9 A0A091HKZ6 F6VXV4 A0A2I0M896 H0Z6W3 A0A3Q3XA48 A0A093Q8R4 A0A0A0A5U5 A0A099ZT44 A0A3S2P745 A0A3L8SD17 A0A1V4JS62 A0A146Y6I4 A0A3Q2NQ52 A0A087RB13 A0A2I4BYH7 A0A147AS80 A0A3P8YJC0 A0A3M0K3H5 A0A093HE96 A0A218V5J8 A0A1A8B005 A0A3Q2Z6U6 A0A091JLX1 H3BG34 A0A1V4JS94 A0A091GXZ8 A0A093NM27 A0A091WM90
A0A139WCL9 K7IXD4 A0A154P067 A0A151XI81 A0A310SPW8 A0A232EV42 W5MD80 H2YNA2 A0A026WAQ2 A0A2A3EHE8 A0A1L8I155 A0A195EQ42 A0A087ZPP4 F7ABS5 A0A3Q1IBX8 A0A3Q0K5M8 V4BRV3 A0A151I7P7 A0A210QD55 F7EE82 A0A3P8W6T1 K1Q7E1 A0A1I8HI71 W5MDA3 A0A267GMP3 R7UDH9 A0A1I8JEL0 A0A060XJ43 A0A267GPP3 B3RYZ2 F1Q8Y1 A0A267DJZ0 A0A2R8RY46 A0A369S1V2 G3VUV1 A0A3B4CV03 A0A3B4ZRR4 C3XUL0 A0A3Q1C600 K7G9M2 A3QK38 A0A151NSZ7 K7G9P0 A0A091NQT4 A0A1U7RLX3 A0A3Q1F350 A0A3B4VE41 F7FU35 A0A3B4Y5N2 A0A093ISJ5 A0A087VDC4 A0A1S3K2G4 A0A2D0PRQ2 A0A1S3K1S6 A0A1I8GK17 A0A3P8UDA3 A0A1W4ZXE4 A0A1X7VIL7 A0A091U632 A0A094KT60 A0A091PNI1 A0A091KE96 M4ASZ2 A0A3Q3BAU9 A0A091SNI9 A0A091HKZ6 F6VXV4 A0A2I0M896 H0Z6W3 A0A3Q3XA48 A0A093Q8R4 A0A0A0A5U5 A0A099ZT44 A0A3S2P745 A0A3L8SD17 A0A1V4JS62 A0A146Y6I4 A0A3Q2NQ52 A0A087RB13 A0A2I4BYH7 A0A147AS80 A0A3P8YJC0 A0A3M0K3H5 A0A093HE96 A0A218V5J8 A0A1A8B005 A0A3Q2Z6U6 A0A091JLX1 H3BG34 A0A1V4JS94 A0A091GXZ8 A0A093NM27 A0A091WM90
Ontologies
GO
Topology
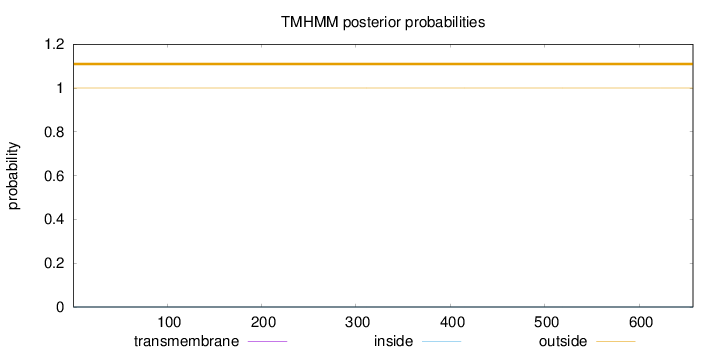
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00399
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.00032
outside
1 - 657
Population Genetic Test Statistics
Pi
22.406115
Theta
24.655891
Tajima's D
-0.647329
CLR
2.42307
CSRT
0.210839458027099
Interpretation
Uncertain
