Gene
KWMTBOMO05994
Pre Gene Modal
BGIBMGA002885
Annotation
PREDICTED:_matrix_metalloproteinase-25-like_[Papilio_machaon]
Full name
Matrix metalloproteinase-25
Alternative Name
Leukolysin
Membrane-type matrix metalloproteinase 6
Membrane-type-6 matrix metalloproteinase
Membrane-type matrix metalloproteinase 6
Membrane-type-6 matrix metalloproteinase
Location in the cell
Cytoplasmic Reliability : 1.967
Sequence
CDS
ATGTTATACCAGAATGCATTGATCACTCAGTTAGCTGCCGCTGAGACTATATCCAAGAATCAATACAAAATATATGACTGTATTCAGTACTCCGACACAATGCTTTCCAGTTACATACTTCAAATTTTGTGCATCAGTTTGGTGCAGTGTAGAACCATTTATTTGGAGCTAAACTCCCCTACGCTTGACGAGGTCAATTTTTTGCTACAATATGGATACTTATCGAAAGATTTGGCCGGAGTTGGCTACACTTACACGTCACAATCTATTTCCGAAGCGGTCAAGAAAATGCAGGCCTTCGCAGGGCTACCACAAACAGGAGTACTGGATGAACCAACTAAGCAGTTATTCAAAAGAAAAAGATGCGGTTTGAAAGATATAGATGAAGACTCGCACGCAAGAAATCGCAGATACATTATACAAGAAGGATGGAATAAACGCGAGGTCACTTACAGACTCCTGAATGGTTCAAGTACAATGAGCAAGGATCGCATAGAAAGGCTTCTAGAGAATGGTTTGGAGGTTTGGGCGCCCCACGGGAACCTCCACTTCACGAAACTAGATGAAGGCAAAGCGGACATACAAGTTTATTTCGCTTCCGGCAACCATGGAGACGGGTTCCCCTTCGATGGCCCTGGTCGAGTAGTGGCTCACGCGTTTCCACCGCCACTCGGCGATATACATTTTGATGATGACGAGACCTGGGGAGTTGAGTCTGATATATCTGAAGACGGAGAAGAAGACATCACGGACTTCTTCGCGGTGGCCGTCCACGAGATCGGACACTCTCTGGGCATGTCACATTCTAACGTCAAATCTTCTGTTATGTATCCATATTATCAGCTGCCCGTTGACAAGCTACACGTGGACGACATATTGGGCATGCAAGAGTTGTACTTAAACGTAAAAGGTTCTGAAGAATCAGAGGGCACCGAACGCACGGTAGGGTCGTCGCAGGCGCCCAGGTTCACGAAGACGGACAGCGAGGAGTTCGATGACGCGCCCGACCTATGCATGACCAACTACGACACCTTGCAAGTCATACACGGCAAAATATACGTGTTCGAAGAAGAGTGGGTCACAGTTCTTCGTGATAGAAACGTCAGAGAGCCCGGGTACCCGAAGAGATTCCATGAGCTTTTCATTGGTCTCCCGTCCAGCATCAAAGTTATACGGACCATCTATGAGAAACACGATCGGAACGTTGTCGTCTTTTCAGAAAACAAATACTGGGAGTTCAGTCCGTCATTCCGTCTGATGAGGCGTGGCTCTCTCACGGATTACTCCATCCCGGACAACGTTACAGAGTTGACCACAGTGTTCATATCCAATTACAACAACAGGACATATCTGATCGAGGAGGAAAGGTATTGGCGGTACGATGAACTAACCGGTACGATGGATGAAGATTACCCAAAGGATATGTCGACATGGCGGGACGTACCGTACCCTGTCGATGCTGCTATTGTTTGGAAAGAAGACACATTCTTCTTCCAAGGTCCTCGTTTTTGGCGCTTTGATAACAACCTCGTCCGAGCACACGAGTACTACCCGCTCCCGACCGCACAGATATGGTTCGACTGCGAATCCAACTTCGAAATGACCTACTACACCATCAACGACGGACCATAA
Protein
MLYQNALITQLAAAETISKNQYKIYDCIQYSDTMLSSYILQILCISLVQCRTIYLELNSPTLDEVNFLLQYGYLSKDLAGVGYTYTSQSISEAVKKMQAFAGLPQTGVLDEPTKQLFKRKRCGLKDIDEDSHARNRRYIIQEGWNKREVTYRLLNGSSTMSKDRIERLLENGLEVWAPHGNLHFTKLDEGKADIQVYFASGNHGDGFPFDGPGRVVAHAFPPPLGDIHFDDDETWGVESDISEDGEEDITDFFAVAVHEIGHSLGMSHSNVKSSVMYPYYQLPVDKLHVDDILGMQELYLNVKGSEESEGTERTVGSSQAPRFTKTDSEEFDDAPDLCMTNYDTLQVIHGKIYVFEEEWVTVLRDRNVREPGYPKRFHELFIGLPSSIKVIRTIYEKHDRNVVVFSENKYWEFSPSFRLMRRGSLTDYSIPDNVTELTTVFISNYNNRTYLIEEERYWRYDELTGTMDEDYPKDMSTWRDVPYPVDAAIVWKEDTFFFQGPRFWRFDNNLVRAHEYYPLPTAQIWFDCESNFEMTYYTINDGP
Summary
Description
May activate progelatinase A.
Cofactor
Zn(2+)
Ca(2+)
Ca(2+)
Miscellaneous
The sequence shown here is derived from an EMBL/GenBank/DDBJ third party annotation (TPA) entry.
Similarity
Belongs to the peptidase M10A family.
Keywords
Calcium
Cell membrane
Cleavage on pair of basic residues
Complete proteome
Disulfide bond
Extracellular matrix
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Membrane
Metal-binding
Metalloprotease
Protease
Reference proteome
Repeat
Secreted
Signal
Zinc
Zymogen
Transmembrane
Transmembrane helix
Feature
chain Matrix metalloproteinase-25
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9J050
A0A2A4JN69
A0A2H1VTH2
A0A3S2MAS1
A0A194QR79
A0A194Q490
+ More
A0A212EHL2 A0A1W4XAM5 A0A0P6IUC8 Q17GF9 T1J720 V4B204 E9H7T7 A0A0V1G433 A0A0V1JCY4 A0A0V0UA13 A0A0V1EZ73 A0A0V1AX01 A0A0V1NVP9 A0A0V1DCQ9 A0A0V0YC80 A0A0V1LUQ3 A0A0V0X6E0 A0A0V1N7S9 A0A0V1AGY6 A0A0V1HWH9 A0A1B6E667 A0A1S3IIW9 A0A2U3YHN6 A0A1B6JEY3 A0A2Y9HQC8 A0A341CK80 A0A340WWZ7 A0A341CMU7 A0A3Q7VEP0 A0A1S3IHV8 M3YUT5 A0A1S3ANG9 A0A2K6LW56 A0A2Y9KLT4 I3LFQ7 A0A2H8TY38 I3ISV9 A0A2Y9PY05 A0A3B4EIB8 A0A2Y9EET4 A0A2U3ZJG9 Q9NPA2 G1REM2 H0XKJ6 A0A1L8EZ04 A0A3Q7PH61 H2QAF3 Q3U435 A0A1S3IH52 F6ZUV1 M5FK30 A0A0B6Z285 A0A1W0XBN1 K1R1S7 A0A210PXF2 A0A1L8HQI6 A0A384A469 A0A085MEC9 A0A2K5N5V4 A0A0D9RCT1 A0A2K5IAT9 Q86GD7 A0A1L8ERA4 A0A3B1K2T9 A0A2K5ZWB8 A0A096NLA4 A0A3P8UVH5 A0A3P8UTX2 A0A2K5UE31 F6YUR1 A0A1L8I0U7 A0A3L7HAA5 F1N1F5 A4IIY9 A0A131XDU2 A0A2K6FVN0 A0A3P8UY86 F6QXB2 M3X751 A0A087YP28 H9GGG5 A0A2K5PYH1 A0A3B3U183 A0A1U7QY82 A0A3P8UY75 T1KNP2 A0A2G9R803 A0A3B3WWI7 A0A2K5E2B0 A0A1S3LFN6 G1U2J8 A0A2I3SF35 A0A2U9CEQ6 A0A2K6U6Z4
A0A212EHL2 A0A1W4XAM5 A0A0P6IUC8 Q17GF9 T1J720 V4B204 E9H7T7 A0A0V1G433 A0A0V1JCY4 A0A0V0UA13 A0A0V1EZ73 A0A0V1AX01 A0A0V1NVP9 A0A0V1DCQ9 A0A0V0YC80 A0A0V1LUQ3 A0A0V0X6E0 A0A0V1N7S9 A0A0V1AGY6 A0A0V1HWH9 A0A1B6E667 A0A1S3IIW9 A0A2U3YHN6 A0A1B6JEY3 A0A2Y9HQC8 A0A341CK80 A0A340WWZ7 A0A341CMU7 A0A3Q7VEP0 A0A1S3IHV8 M3YUT5 A0A1S3ANG9 A0A2K6LW56 A0A2Y9KLT4 I3LFQ7 A0A2H8TY38 I3ISV9 A0A2Y9PY05 A0A3B4EIB8 A0A2Y9EET4 A0A2U3ZJG9 Q9NPA2 G1REM2 H0XKJ6 A0A1L8EZ04 A0A3Q7PH61 H2QAF3 Q3U435 A0A1S3IH52 F6ZUV1 M5FK30 A0A0B6Z285 A0A1W0XBN1 K1R1S7 A0A210PXF2 A0A1L8HQI6 A0A384A469 A0A085MEC9 A0A2K5N5V4 A0A0D9RCT1 A0A2K5IAT9 Q86GD7 A0A1L8ERA4 A0A3B1K2T9 A0A2K5ZWB8 A0A096NLA4 A0A3P8UVH5 A0A3P8UTX2 A0A2K5UE31 F6YUR1 A0A1L8I0U7 A0A3L7HAA5 F1N1F5 A4IIY9 A0A131XDU2 A0A2K6FVN0 A0A3P8UY86 F6QXB2 M3X751 A0A087YP28 H9GGG5 A0A2K5PYH1 A0A3B3U183 A0A1U7QY82 A0A3P8UY75 T1KNP2 A0A2G9R803 A0A3B3WWI7 A0A2K5E2B0 A0A1S3LFN6 G1U2J8 A0A2I3SF35 A0A2U9CEQ6 A0A2K6U6Z4
EC Number
3.4.24.-
Pubmed
EMBL
BABH01018950
BABH01018951
BABH01018952
NWSH01001058
PCG72883.1
ODYU01004342
+ More
SOQ44113.1 RSAL01000002 RVE54959.1 KQ461175 KPJ07992.1 KQ459472 KPJ00164.1 AGBW02014864 OWR40976.1 GDUN01000558 JAN95361.1 CH477259 EAT45766.1 JH431902 KB200505 ESP01496.1 GL732601 EFX72236.1 JYDT01000004 KRY92957.1 JYDS01000013 JYDV01000001 KRZ32835.1 KRZ46247.1 JYDJ01000035 KRX47973.1 JYDR01000003 KRY78876.1 JYDH01000192 KRY28755.1 JYDM01000092 KRZ87979.1 JYDI01000013 KRY59303.1 JYDU01000025 KRX98046.1 JYDW01000003 KRZ63153.1 JYDK01000012 KRX83564.1 JYDO01000004 KRZ80010.1 JYDQ01000001 KRY23933.1 JYDP01000022 KRZ14720.1 GEDC01025185 GEDC01003893 JAS12113.1 JAS33405.1 GECU01009845 JAS97861.1 AEYP01027796 AEYP01027797 AEYP01027798 AEMK02000018 GFXV01006637 MBW18442.1 BX510645 LO018620 AJ239053 AF145442 AF185270 AB042328 AJ272137 CH471112 ADFV01048390 ADFV01048391 AAQR03078194 AAQR03078195 AAQR03078196 CM004482 OCT64582.1 AC196449 NBAG03000299 PNI44531.1 AK154458 BC112379 GJ062847 DAA15654.1 HACG01015176 CEK62041.1 MTYJ01000005 OQV24830.1 JH818024 EKC39793.1 NEDP02005420 OWF41146.1 CM004467 OCT98341.1 KL363199 KFD55575.1 AQIB01117902 AB106142 BAC66058.1 CM004483 OCT61861.1 AHZZ02018218 AQIA01041731 JSUE03025506 CM004466 OCU01955.1 RAZU01000243 RLQ62888.1 BC136206 AAI36207.1 GEFH01004800 JAP63781.1 AAMC01006081 AAMC01006082 AAMC01006083 AANG04001668 AYCK01001016 AYCK01001017 AYCK01001018 AYCK01001019 AYCK01001020 AAWZ02029520 AAWZ02029521 AAWZ02029522 AAWZ02029523 CAEY01000277 KV955468 PIO23990.1 CP026258 AWP15074.1
SOQ44113.1 RSAL01000002 RVE54959.1 KQ461175 KPJ07992.1 KQ459472 KPJ00164.1 AGBW02014864 OWR40976.1 GDUN01000558 JAN95361.1 CH477259 EAT45766.1 JH431902 KB200505 ESP01496.1 GL732601 EFX72236.1 JYDT01000004 KRY92957.1 JYDS01000013 JYDV01000001 KRZ32835.1 KRZ46247.1 JYDJ01000035 KRX47973.1 JYDR01000003 KRY78876.1 JYDH01000192 KRY28755.1 JYDM01000092 KRZ87979.1 JYDI01000013 KRY59303.1 JYDU01000025 KRX98046.1 JYDW01000003 KRZ63153.1 JYDK01000012 KRX83564.1 JYDO01000004 KRZ80010.1 JYDQ01000001 KRY23933.1 JYDP01000022 KRZ14720.1 GEDC01025185 GEDC01003893 JAS12113.1 JAS33405.1 GECU01009845 JAS97861.1 AEYP01027796 AEYP01027797 AEYP01027798 AEMK02000018 GFXV01006637 MBW18442.1 BX510645 LO018620 AJ239053 AF145442 AF185270 AB042328 AJ272137 CH471112 ADFV01048390 ADFV01048391 AAQR03078194 AAQR03078195 AAQR03078196 CM004482 OCT64582.1 AC196449 NBAG03000299 PNI44531.1 AK154458 BC112379 GJ062847 DAA15654.1 HACG01015176 CEK62041.1 MTYJ01000005 OQV24830.1 JH818024 EKC39793.1 NEDP02005420 OWF41146.1 CM004467 OCT98341.1 KL363199 KFD55575.1 AQIB01117902 AB106142 BAC66058.1 CM004483 OCT61861.1 AHZZ02018218 AQIA01041731 JSUE03025506 CM004466 OCU01955.1 RAZU01000243 RLQ62888.1 BC136206 AAI36207.1 GEFH01004800 JAP63781.1 AAMC01006081 AAMC01006082 AAMC01006083 AANG04001668 AYCK01001016 AYCK01001017 AYCK01001018 AYCK01001019 AYCK01001020 AAWZ02029520 AAWZ02029521 AAWZ02029522 AAWZ02029523 CAEY01000277 KV955468 PIO23990.1 CP026258 AWP15074.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000192223 UP000008820 UP000030746 UP000000305 UP000054995 UP000054805 UP000054826 UP000055048 UP000054632 UP000054776 UP000054924 UP000054653 UP000054815 UP000054721 UP000054673 UP000054843 UP000054783 UP000055024 UP000085678 UP000245341 UP000248481 UP000252040 UP000265300 UP000286642 UP000000715 UP000079721 UP000233180 UP000248482 UP000008227 UP000000437 UP000248483 UP000261440 UP000248484 UP000245340 UP000005640 UP000001073 UP000005225 UP000186698 UP000286641 UP000002277 UP000000589 UP000002281 UP000005408 UP000242188 UP000261681 UP000233060 UP000029965 UP000233080 UP000018467 UP000233140 UP000028761 UP000265120 UP000233100 UP000006718 UP000273346 UP000009136 UP000233160 UP000008143 UP000011712 UP000028760 UP000001646 UP000233040 UP000261500 UP000189706 UP000015104 UP000261480 UP000233020 UP000087266 UP000001811 UP000246464 UP000233220
UP000192223 UP000008820 UP000030746 UP000000305 UP000054995 UP000054805 UP000054826 UP000055048 UP000054632 UP000054776 UP000054924 UP000054653 UP000054815 UP000054721 UP000054673 UP000054843 UP000054783 UP000055024 UP000085678 UP000245341 UP000248481 UP000252040 UP000265300 UP000286642 UP000000715 UP000079721 UP000233180 UP000248482 UP000008227 UP000000437 UP000248483 UP000261440 UP000248484 UP000245340 UP000005640 UP000001073 UP000005225 UP000186698 UP000286641 UP000002277 UP000000589 UP000002281 UP000005408 UP000242188 UP000261681 UP000233060 UP000029965 UP000233080 UP000018467 UP000233140 UP000028761 UP000265120 UP000233100 UP000006718 UP000273346 UP000009136 UP000233160 UP000008143 UP000011712 UP000028760 UP000001646 UP000233040 UP000261500 UP000189706 UP000015104 UP000261480 UP000233020 UP000087266 UP000001811 UP000246464 UP000233220
Interpro
IPR006026
Peptidase_Metallo
+ More
IPR018487 Hemopexin-like_repeat
IPR024079 MetalloPept_cat_dom_sf
IPR021190 Pept_M10A
IPR033739 M10A_MMP
IPR036375 Hemopexin-like_dom_sf
IPR001818 Pept_M10_metallopeptidase
IPR028733 MMP25
IPR002477 Peptidoglycan-bd-like
IPR036365 PGBD-like_sf
IPR000585 Hemopexin-like_dom
IPR021158 Pept_M10A_Zn_BS
IPR028726 MMP17
IPR018486 Hemopexin_CS
IPR018487 Hemopexin-like_repeat
IPR024079 MetalloPept_cat_dom_sf
IPR021190 Pept_M10A
IPR033739 M10A_MMP
IPR036375 Hemopexin-like_dom_sf
IPR001818 Pept_M10_metallopeptidase
IPR028733 MMP25
IPR002477 Peptidoglycan-bd-like
IPR036365 PGBD-like_sf
IPR000585 Hemopexin-like_dom
IPR021158 Pept_M10A_Zn_BS
IPR028726 MMP17
IPR018486 Hemopexin_CS
Gene 3D
ProteinModelPortal
H9J050
A0A2A4JN69
A0A2H1VTH2
A0A3S2MAS1
A0A194QR79
A0A194Q490
+ More
A0A212EHL2 A0A1W4XAM5 A0A0P6IUC8 Q17GF9 T1J720 V4B204 E9H7T7 A0A0V1G433 A0A0V1JCY4 A0A0V0UA13 A0A0V1EZ73 A0A0V1AX01 A0A0V1NVP9 A0A0V1DCQ9 A0A0V0YC80 A0A0V1LUQ3 A0A0V0X6E0 A0A0V1N7S9 A0A0V1AGY6 A0A0V1HWH9 A0A1B6E667 A0A1S3IIW9 A0A2U3YHN6 A0A1B6JEY3 A0A2Y9HQC8 A0A341CK80 A0A340WWZ7 A0A341CMU7 A0A3Q7VEP0 A0A1S3IHV8 M3YUT5 A0A1S3ANG9 A0A2K6LW56 A0A2Y9KLT4 I3LFQ7 A0A2H8TY38 I3ISV9 A0A2Y9PY05 A0A3B4EIB8 A0A2Y9EET4 A0A2U3ZJG9 Q9NPA2 G1REM2 H0XKJ6 A0A1L8EZ04 A0A3Q7PH61 H2QAF3 Q3U435 A0A1S3IH52 F6ZUV1 M5FK30 A0A0B6Z285 A0A1W0XBN1 K1R1S7 A0A210PXF2 A0A1L8HQI6 A0A384A469 A0A085MEC9 A0A2K5N5V4 A0A0D9RCT1 A0A2K5IAT9 Q86GD7 A0A1L8ERA4 A0A3B1K2T9 A0A2K5ZWB8 A0A096NLA4 A0A3P8UVH5 A0A3P8UTX2 A0A2K5UE31 F6YUR1 A0A1L8I0U7 A0A3L7HAA5 F1N1F5 A4IIY9 A0A131XDU2 A0A2K6FVN0 A0A3P8UY86 F6QXB2 M3X751 A0A087YP28 H9GGG5 A0A2K5PYH1 A0A3B3U183 A0A1U7QY82 A0A3P8UY75 T1KNP2 A0A2G9R803 A0A3B3WWI7 A0A2K5E2B0 A0A1S3LFN6 G1U2J8 A0A2I3SF35 A0A2U9CEQ6 A0A2K6U6Z4
A0A212EHL2 A0A1W4XAM5 A0A0P6IUC8 Q17GF9 T1J720 V4B204 E9H7T7 A0A0V1G433 A0A0V1JCY4 A0A0V0UA13 A0A0V1EZ73 A0A0V1AX01 A0A0V1NVP9 A0A0V1DCQ9 A0A0V0YC80 A0A0V1LUQ3 A0A0V0X6E0 A0A0V1N7S9 A0A0V1AGY6 A0A0V1HWH9 A0A1B6E667 A0A1S3IIW9 A0A2U3YHN6 A0A1B6JEY3 A0A2Y9HQC8 A0A341CK80 A0A340WWZ7 A0A341CMU7 A0A3Q7VEP0 A0A1S3IHV8 M3YUT5 A0A1S3ANG9 A0A2K6LW56 A0A2Y9KLT4 I3LFQ7 A0A2H8TY38 I3ISV9 A0A2Y9PY05 A0A3B4EIB8 A0A2Y9EET4 A0A2U3ZJG9 Q9NPA2 G1REM2 H0XKJ6 A0A1L8EZ04 A0A3Q7PH61 H2QAF3 Q3U435 A0A1S3IH52 F6ZUV1 M5FK30 A0A0B6Z285 A0A1W0XBN1 K1R1S7 A0A210PXF2 A0A1L8HQI6 A0A384A469 A0A085MEC9 A0A2K5N5V4 A0A0D9RCT1 A0A2K5IAT9 Q86GD7 A0A1L8ERA4 A0A3B1K2T9 A0A2K5ZWB8 A0A096NLA4 A0A3P8UVH5 A0A3P8UTX2 A0A2K5UE31 F6YUR1 A0A1L8I0U7 A0A3L7HAA5 F1N1F5 A4IIY9 A0A131XDU2 A0A2K6FVN0 A0A3P8UY86 F6QXB2 M3X751 A0A087YP28 H9GGG5 A0A2K5PYH1 A0A3B3U183 A0A1U7QY82 A0A3P8UY75 T1KNP2 A0A2G9R803 A0A3B3WWI7 A0A2K5E2B0 A0A1S3LFN6 G1U2J8 A0A2I3SF35 A0A2U9CEQ6 A0A2K6U6Z4
PDB
1SU3
E-value=1.28728e-48,
Score=489
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Secreted
Extracellular space
Extracellular matrix
Secreted
Extracellular space
Extracellular matrix
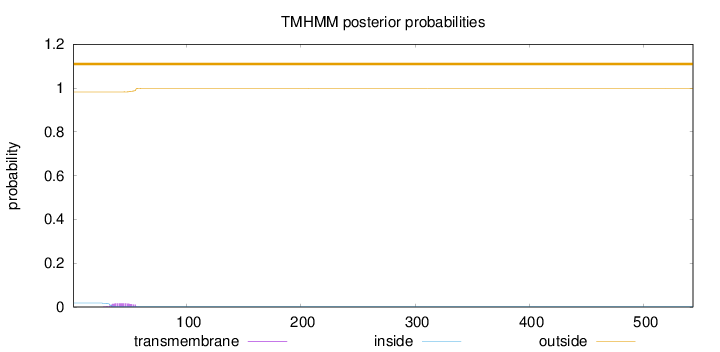
Length:
543
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.353150000000001
Exp number, first 60 AAs:
0.35092
Total prob of N-in:
0.01781
outside
1 - 543
Population Genetic Test Statistics
Pi
16.009646
Theta
20.148202
Tajima's D
-1.048133
CLR
10.012875
CSRT
0.128343582820859
Interpretation
Uncertain
