Gene
KWMTBOMO05993 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002818
Annotation
PREDICTED:_thioredoxin_reductase_1?_mitochondrial_isoform_X2_[Bombyx_mori]
Full name
Thioredoxin reductase 1, mitochondrial
Location in the cell
Cytoplasmic Reliability : 2.58
Sequence
CDS
ATGGCACCTGTCGCAGGAACATACGACTATGATCTGGCTGTAATCGGAGGTGGCTCCGGTGGACTGGCATGCGCCAAGGAGGCTGTGAACTTGGGTGCTAAAGTGACTGTGTTGGACTACGTCACACCCTCACCGCAAGGCACCAAGTGGGGCCTGGGAGGAACCTGTGTTAATGTAGGATGTATTCCGAAGAAGCTGATGCATCAGGCTGCTTTACTTGGAGAGAGCATACATGAAGCGGTTGCATACGGTTGGGAGGTGCCCTCACTGGACGCGATCAAGATAAACTGGCCAGCGCTAACTGAAGCTGTACAGAATCACATAAAATCTGTGAACTGGGTGACCAGGGTTGACCTGAGAGAGAAGAAAATAGATTACGTTAACGGTCTGGGTGAGTTTAAGGACGCCCACACGCTGATAGCGACCTTGAAGAATGGATCGAAGAAGGAGATCACGGCTAAGAATATTGTGATCGCCGTGGGCGGTAGGCCGCACTACCCGGACATTCCTGGGGCGGTCGAGTACTGCATCAGCAGTGACGATATCTTCAGTTTAGGTCATCCCCCCGGGAAGACCCTGGTCGTTGGAGCTGGATACATCGGCTTAGAATGCGCCGGATTTTTGAACTCTTTGGGTTACCCGGCCACCGTGCTGGTCCGATCGGTGCCACTCCGCGGCTTCGACCAGCAAATGGCCCAGGCGGTAACCAGCGAGATGGAACAGAAGGGAGTCGTATTCCACAACAAGTGCGTACCGCTGTCCGTGGAGAAGCTGGAGACCGGTCAGTTGAAGGCGCGCTGGCAGAACACGGAGACCCAAGAGCGGGGTGAGGATGTGTTCGATACGGTGCTGATGGCGACCGGCCGGTACGCCCTCACCAAAACCCTCAACCTCGAAGCGGCAGGAGTGACGTGTGTATCAAACAGCGGGAAGATCATAGCTGAAACTGAACAGACCAACGTCTCTAACATATACGCGGTGGGCGACGTGTTGGAGGGGAAGCCTGAGCTGACCCCCGTGGCCATCCACGCGGGCCGGCTGCTGGCGCGCCGGATGTTCGCCGGGGCCACGCAGCCCATGGACTACGACAACGTGGCCACCACCGTGTTCACGCCGTTAGAGTACGGCTGCGTCGGGCTAAGCGAGGAGACCGCCCTGGCCAGGCACGGCGCCGACAAGGTGGAGGTGTACCACGCGTTCTACAAACCGACGGAGTTCTTCATACCGCAGAGGAACATCCGCAACTGCTACCTGAAGGCCGTGGCGCTCCGCGAGGCGCCGCAGCGCATCCTCGGGCTCCACTTTGTGGGGCCGGTCGCCGGCGAGGTCATACAGGGCTTCGCAGCTGCCGTCAAATGCGGCATAACTATGGAGCAATTGATGAACACAGTCGGCATTCACCCCACGGTGGCCGAGGAGTTCACCAGACTCAACATCACCAAGCGCTCCGGCAAGGACCCCAACCCTGCGTCGTGCTGCAGCTAG
Protein
MAPVAGTYDYDLAVIGGGSGGLACAKEAVNLGAKVTVLDYVTPSPQGTKWGLGGTCVNVGCIPKKLMHQAALLGESIHEAVAYGWEVPSLDAIKINWPALTEAVQNHIKSVNWVTRVDLREKKIDYVNGLGEFKDAHTLIATLKNGSKKEITAKNIVIAVGGRPHYPDIPGAVEYCISSDDIFSLGHPPGKTLVVGAGYIGLECAGFLNSLGYPATVLVRSVPLRGFDQQMAQAVTSEMEQKGVVFHNKCVPLSVEKLETGQLKARWQNTETQERGEDVFDTVLMATGRYALTKTLNLEAAGVTCVSNSGKIIAETEQTNVSNIYAVGDVLEGKPELTPVAIHAGRLLARRMFAGATQPMDYDNVATTVFTPLEYGCVGLSEETALARHGADKVEVYHAFYKPTEFFIPQRNIRNCYLKAVALREAPQRILGLHFVGPVAGEVIQGFAAAVKCGITMEQLMNTVGIHPTVAEEFTRLNITKRSGKDPNPASCCS
Summary
Description
Thioredoxin system is a major player in glutathione metabolism, due to the demonstrated absence of a glutathione reductase. Functionally interacts with the Sod/Cat reactive oxidation species (ROS) defense system and thereby has a role in preadult development and life span. Lack of a glutathione reductase suggests antioxidant defense in Drosophila, and probably in related insects, differs fundamentally from that in other organisms.
Catalytic Activity
[thioredoxin]-dithiol + NADP(+) = [thioredoxin]-disulfide + H(+) + NADPH
Cofactor
FAD
Biophysicochemical Properties
6.5 uM for NADPH (at pH 7.4)
1 uM for NADPH (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
1 uM for NADPH (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
7.0 uM for dhd (at pH 7.4, 200 uM NADPH, 100 mM KPO(4))
141 uM for dhd (at pH 7.0, 0.15 mM NADPH, 1 mM EDTA, 1 mg/ml insulin, 50 mM KPO(4), 25 degrees Celsius)
7 uM for dhd (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
19 uM for dhd (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
5.9 uM for Trx-2 (at pH 7.4, 100 uM NADPH, 2 mM EDTA, 100 mM KPO(4))
310 uM for 5,5'-dithiobis-2-nitrobenzoic acid (DTNB) (at pH 7.4, 100 uM NADPH, 100 mM KPO(4))
0.17 mM for DTNB (at pH 7.0, 0.2 mM NADPH, 10 mM EDTA, 100 mM KPO(4), 25 degrees Celsius)
380 uM for DTNB (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
410 uM for DTNB (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
675 uM for methylseleninate (100 uM NADPH)
1 uM for NADPH (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
1 uM for NADPH (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
7.0 uM for dhd (at pH 7.4, 200 uM NADPH, 100 mM KPO(4))
141 uM for dhd (at pH 7.0, 0.15 mM NADPH, 1 mM EDTA, 1 mg/ml insulin, 50 mM KPO(4), 25 degrees Celsius)
7 uM for dhd (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
19 uM for dhd (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
5.9 uM for Trx-2 (at pH 7.4, 100 uM NADPH, 2 mM EDTA, 100 mM KPO(4))
310 uM for 5,5'-dithiobis-2-nitrobenzoic acid (DTNB) (at pH 7.4, 100 uM NADPH, 100 mM KPO(4))
0.17 mM for DTNB (at pH 7.0, 0.2 mM NADPH, 10 mM EDTA, 100 mM KPO(4), 25 degrees Celsius)
380 uM for DTNB (at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
410 uM for DTNB (isoform B at pH 7.4, 2 mM EDTA, 100 mM KPO(4))
675 uM for methylseleninate (100 uM NADPH)
Subunit
Homodimer.
Miscellaneous
The active site is a redox-active disulfide bond.
Similarity
Belongs to the class-I pyridine nucleotide-disulfide oxidoreductase family.
Keywords
3D-structure
Alternative initiation
Alternative splicing
Complete proteome
Cytoplasm
Disulfide bond
FAD
Flavoprotein
Mitochondrion
NADP
Nucleotide-binding
Oxidoreductase
Redox-active center
Reference proteome
Transit peptide
Feature
chain Thioredoxin reductase 1, mitochondrial
splice variant In isoform A and isoform D.
splice variant In isoform A and isoform D.
Uniprot
H9IZY3
S4PGR5
A0A2H1VTD3
A0A0A7RB97
A0A194QSN1
A0A3S2LW17
+ More
A0A212EHM0 A0A2A4JMR9 W8BVK8 A0A1L8EFX8 A0A0A1WJ37 A0A034VC07 A0A1L8DZH0 A0A1L8EFM1 A0A1I8P8B7 A0A1I8P8E9 A0A2J7PD15 A0A1I8P889 A0A2J7PD09 A0A1L8E9N4 T1PE19 A0A034V6Y5 P91884 A0A034VAF2 A0A1I8P866 W5JC17 A0A182Y7I3 A0A336MJC3 A0A336L8M5 A0A1L8DZG2 A0A3B0KKV3 A0A2M4CIZ1 A0A1W4XAK5 A0A1W4X9D2 Q29FR6 A0A2M4CHN3 A0A2M4CHN1 A0A2M4CHX9 A0A2M3Z590 W5JM65 A0A182PQ49 A0A182RV39 J3JWU4 A0A084VFN0 Q86GB3 A0A2M4AB12 A0A2M4A9Z4 Q86GB2 A0A0N8P0L5 Q8MZM6 A0A0R3P666 A0A2M4AAC2 A0A2M4BJZ2 A0A182UQM2 A0A067RB75 A0A182IA68 A0A182FJY4 A0A2M4BK59 A0A1W4VX70 A0A2M4ADP1 A0A182XLQ9 A0A182NVI1 A0A182W4R7 U5EQ42 A0A2M4AD74 A0A182F127 A0A182MCK5 A0A182QER2 A0A182U8H8 A0A2M4BL19 B4Q0D5 A0A182KEH4 A0A2M4ABX6 B3NX17 B3MXU2 A0A0L0BM47 A0A0Q9WU01 A0A2M3ZGC9 A0A2M4AIK6 A0A2M3Z2G9 A0A0K8TUD9 A0A1W4VJ61 P91938-2 A0A1W4VWE3 A0A0R1EHY5 A0A0R1EBL0 A0A1J1IYM2 A0A1B6CAT4 A0A0P4VIW8 A0A0Q5T6X3 D6WDX1 B4NDH3 A0A0Q5TAX5 A0A1B6CT38 A0A0V0G4M4 A0A182J4F6 A0A1B6BXA1 A0A069DV52 A0A1A9WL68 Q17GT5
A0A212EHM0 A0A2A4JMR9 W8BVK8 A0A1L8EFX8 A0A0A1WJ37 A0A034VC07 A0A1L8DZH0 A0A1L8EFM1 A0A1I8P8B7 A0A1I8P8E9 A0A2J7PD15 A0A1I8P889 A0A2J7PD09 A0A1L8E9N4 T1PE19 A0A034V6Y5 P91884 A0A034VAF2 A0A1I8P866 W5JC17 A0A182Y7I3 A0A336MJC3 A0A336L8M5 A0A1L8DZG2 A0A3B0KKV3 A0A2M4CIZ1 A0A1W4XAK5 A0A1W4X9D2 Q29FR6 A0A2M4CHN3 A0A2M4CHN1 A0A2M4CHX9 A0A2M3Z590 W5JM65 A0A182PQ49 A0A182RV39 J3JWU4 A0A084VFN0 Q86GB3 A0A2M4AB12 A0A2M4A9Z4 Q86GB2 A0A0N8P0L5 Q8MZM6 A0A0R3P666 A0A2M4AAC2 A0A2M4BJZ2 A0A182UQM2 A0A067RB75 A0A182IA68 A0A182FJY4 A0A2M4BK59 A0A1W4VX70 A0A2M4ADP1 A0A182XLQ9 A0A182NVI1 A0A182W4R7 U5EQ42 A0A2M4AD74 A0A182F127 A0A182MCK5 A0A182QER2 A0A182U8H8 A0A2M4BL19 B4Q0D5 A0A182KEH4 A0A2M4ABX6 B3NX17 B3MXU2 A0A0L0BM47 A0A0Q9WU01 A0A2M3ZGC9 A0A2M4AIK6 A0A2M3Z2G9 A0A0K8TUD9 A0A1W4VJ61 P91938-2 A0A1W4VWE3 A0A0R1EHY5 A0A0R1EBL0 A0A1J1IYM2 A0A1B6CAT4 A0A0P4VIW8 A0A0Q5T6X3 D6WDX1 B4NDH3 A0A0Q5TAX5 A0A1B6CT38 A0A0V0G4M4 A0A182J4F6 A0A1B6BXA1 A0A069DV52 A0A1A9WL68 Q17GT5
EC Number
1.8.1.9
Pubmed
19121390
23622113
26502992
26354079
22118469
24495485
+ More
25830018 25348373 25315136 20920257 23761445 25244985 15632085 17994087 22516182 23537049 24438588 12364791 14747013 17210077 24845553 17550304 26108605 26369729 9056265 11158675 10731132 12537572 12537569 11525742 11782468 11796729 11877442 18211101 18991392 17385893 27129103 18362917 19820115 26334808 17510324
25830018 25348373 25315136 20920257 23761445 25244985 15632085 17994087 22516182 23537049 24438588 12364791 14747013 17210077 24845553 17550304 26108605 26369729 9056265 11158675 10731132 12537572 12537569 11525742 11782468 11796729 11877442 18211101 18991392 17385893 27129103 18362917 19820115 26334808 17510324
EMBL
BABH01018954
GAIX01003537
JAA89023.1
ODYU01004342
SOQ44111.1
KM658552
+ More
AJA32127.1 KQ461175 KPJ07990.1 RSAL01000002 RVE54960.1 AGBW02014864 OWR40977.1 NWSH01001058 PCG72884.1 GAMC01005604 GAMC01005602 JAC00954.1 GFDG01001316 JAV17483.1 GBXI01015894 JAC98397.1 GAKP01019864 GAKP01019863 JAC39088.1 GFDF01002258 JAV11826.1 GFDG01001321 JAV17478.1 NEVH01026394 PNF14225.1 PNF14223.1 GFDG01003395 JAV15404.1 KA646966 AFP61595.1 GAKP01019866 JAC39086.1 U88187 AAC69637.1 GAKP01019865 JAC39087.1 ADMH02001645 ETN61621.1 UFQT01001105 SSX28933.1 UFQS01002506 UFQT01002506 SSX14186.1 SSX33602.1 GFDF01002260 JAV11824.1 OUUW01000018 SPP89210.1 GGFL01000670 MBW64848.1 CH379065 EAL31513.2 GGFL01000672 MBW64850.1 GGFL01000669 MBW64847.1 GGFL01000671 MBW64849.1 GGFM01002929 MBW23680.1 ADMH02000725 ETN65241.1 APGK01058933 BT127712 KB741292 AEE62674.1 ENN70347.1 ATLV01012450 KE524793 KFB36774.1 AJ549084 AAAB01008846 CAD70158.1 EAA06298.3 GGFK01004655 MBW37976.1 GGFK01004286 MBW37607.1 AJ549085 CAD70159.1 EAU77244.1 CH902630 KPU77460.1 AJ459821 CAD30858.1 EGK96416.1 KRT07247.1 GGFK01004433 MBW37754.1 GGFJ01004239 MBW53380.1 KK852620 KDR20063.1 APCN01001127 GGFJ01004271 MBW53412.1 GGFK01005585 MBW38906.1 GANO01003434 JAB56437.1 GGFK01005410 MBW38731.1 AXCM01000581 AXCN02002153 GGFJ01004626 MBW53767.1 CM000162 EDX02272.1 GGFK01004939 MBW38260.1 CH954180 EDV46846.1 EDV38557.2 JRES01001679 KNC20998.1 CH964239 KRF99678.1 GGFM01006876 MBW27627.1 GGFK01007292 MBW40613.1 GGFM01001968 MBW22719.1 GDAI01000048 JAI17555.1 U81995 AF301145 AF301144 AE014298 BT003266 BT025070 AY051643 KRK06682.1 KRK06681.1 CVRI01000063 CRL04794.1 GEDC01026784 JAS10514.1 GDKW01002590 JAI54005.1 KQS30144.1 KQ971323 EEZ99907.2 EDW82879.1 KQS30145.1 GEDC01020741 GEDC01000988 JAS16557.1 JAS36310.1 GECL01003116 JAP03008.1 GEDC01031386 JAS05912.1 GBGD01001193 JAC87696.1 CH477256 EAT45853.1
AJA32127.1 KQ461175 KPJ07990.1 RSAL01000002 RVE54960.1 AGBW02014864 OWR40977.1 NWSH01001058 PCG72884.1 GAMC01005604 GAMC01005602 JAC00954.1 GFDG01001316 JAV17483.1 GBXI01015894 JAC98397.1 GAKP01019864 GAKP01019863 JAC39088.1 GFDF01002258 JAV11826.1 GFDG01001321 JAV17478.1 NEVH01026394 PNF14225.1 PNF14223.1 GFDG01003395 JAV15404.1 KA646966 AFP61595.1 GAKP01019866 JAC39086.1 U88187 AAC69637.1 GAKP01019865 JAC39087.1 ADMH02001645 ETN61621.1 UFQT01001105 SSX28933.1 UFQS01002506 UFQT01002506 SSX14186.1 SSX33602.1 GFDF01002260 JAV11824.1 OUUW01000018 SPP89210.1 GGFL01000670 MBW64848.1 CH379065 EAL31513.2 GGFL01000672 MBW64850.1 GGFL01000669 MBW64847.1 GGFL01000671 MBW64849.1 GGFM01002929 MBW23680.1 ADMH02000725 ETN65241.1 APGK01058933 BT127712 KB741292 AEE62674.1 ENN70347.1 ATLV01012450 KE524793 KFB36774.1 AJ549084 AAAB01008846 CAD70158.1 EAA06298.3 GGFK01004655 MBW37976.1 GGFK01004286 MBW37607.1 AJ549085 CAD70159.1 EAU77244.1 CH902630 KPU77460.1 AJ459821 CAD30858.1 EGK96416.1 KRT07247.1 GGFK01004433 MBW37754.1 GGFJ01004239 MBW53380.1 KK852620 KDR20063.1 APCN01001127 GGFJ01004271 MBW53412.1 GGFK01005585 MBW38906.1 GANO01003434 JAB56437.1 GGFK01005410 MBW38731.1 AXCM01000581 AXCN02002153 GGFJ01004626 MBW53767.1 CM000162 EDX02272.1 GGFK01004939 MBW38260.1 CH954180 EDV46846.1 EDV38557.2 JRES01001679 KNC20998.1 CH964239 KRF99678.1 GGFM01006876 MBW27627.1 GGFK01007292 MBW40613.1 GGFM01001968 MBW22719.1 GDAI01000048 JAI17555.1 U81995 AF301145 AF301144 AE014298 BT003266 BT025070 AY051643 KRK06682.1 KRK06681.1 CVRI01000063 CRL04794.1 GEDC01026784 JAS10514.1 GDKW01002590 JAI54005.1 KQS30144.1 KQ971323 EEZ99907.2 EDW82879.1 KQS30145.1 GEDC01020741 GEDC01000988 JAS16557.1 JAS36310.1 GECL01003116 JAP03008.1 GEDC01031386 JAS05912.1 GBGD01001193 JAC87696.1 CH477256 EAT45853.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000007151
UP000218220
UP000095300
+ More
UP000235965 UP000095301 UP000000673 UP000076408 UP000268350 UP000192223 UP000001819 UP000075885 UP000075900 UP000019118 UP000030765 UP000007062 UP000007801 UP000075903 UP000027135 UP000075840 UP000069272 UP000192221 UP000076407 UP000075884 UP000075920 UP000075883 UP000075886 UP000075902 UP000002282 UP000075881 UP000008711 UP000037069 UP000007798 UP000000803 UP000183832 UP000007266 UP000075880 UP000091820 UP000008820
UP000235965 UP000095301 UP000000673 UP000076408 UP000268350 UP000192223 UP000001819 UP000075885 UP000075900 UP000019118 UP000030765 UP000007062 UP000007801 UP000075903 UP000027135 UP000075840 UP000069272 UP000192221 UP000076407 UP000075884 UP000075920 UP000075883 UP000075886 UP000075902 UP000002282 UP000075881 UP000008711 UP000037069 UP000007798 UP000000803 UP000183832 UP000007266 UP000075880 UP000091820 UP000008820
Interpro
Gene 3D
ProteinModelPortal
H9IZY3
S4PGR5
A0A2H1VTD3
A0A0A7RB97
A0A194QSN1
A0A3S2LW17
+ More
A0A212EHM0 A0A2A4JMR9 W8BVK8 A0A1L8EFX8 A0A0A1WJ37 A0A034VC07 A0A1L8DZH0 A0A1L8EFM1 A0A1I8P8B7 A0A1I8P8E9 A0A2J7PD15 A0A1I8P889 A0A2J7PD09 A0A1L8E9N4 T1PE19 A0A034V6Y5 P91884 A0A034VAF2 A0A1I8P866 W5JC17 A0A182Y7I3 A0A336MJC3 A0A336L8M5 A0A1L8DZG2 A0A3B0KKV3 A0A2M4CIZ1 A0A1W4XAK5 A0A1W4X9D2 Q29FR6 A0A2M4CHN3 A0A2M4CHN1 A0A2M4CHX9 A0A2M3Z590 W5JM65 A0A182PQ49 A0A182RV39 J3JWU4 A0A084VFN0 Q86GB3 A0A2M4AB12 A0A2M4A9Z4 Q86GB2 A0A0N8P0L5 Q8MZM6 A0A0R3P666 A0A2M4AAC2 A0A2M4BJZ2 A0A182UQM2 A0A067RB75 A0A182IA68 A0A182FJY4 A0A2M4BK59 A0A1W4VX70 A0A2M4ADP1 A0A182XLQ9 A0A182NVI1 A0A182W4R7 U5EQ42 A0A2M4AD74 A0A182F127 A0A182MCK5 A0A182QER2 A0A182U8H8 A0A2M4BL19 B4Q0D5 A0A182KEH4 A0A2M4ABX6 B3NX17 B3MXU2 A0A0L0BM47 A0A0Q9WU01 A0A2M3ZGC9 A0A2M4AIK6 A0A2M3Z2G9 A0A0K8TUD9 A0A1W4VJ61 P91938-2 A0A1W4VWE3 A0A0R1EHY5 A0A0R1EBL0 A0A1J1IYM2 A0A1B6CAT4 A0A0P4VIW8 A0A0Q5T6X3 D6WDX1 B4NDH3 A0A0Q5TAX5 A0A1B6CT38 A0A0V0G4M4 A0A182J4F6 A0A1B6BXA1 A0A069DV52 A0A1A9WL68 Q17GT5
A0A212EHM0 A0A2A4JMR9 W8BVK8 A0A1L8EFX8 A0A0A1WJ37 A0A034VC07 A0A1L8DZH0 A0A1L8EFM1 A0A1I8P8B7 A0A1I8P8E9 A0A2J7PD15 A0A1I8P889 A0A2J7PD09 A0A1L8E9N4 T1PE19 A0A034V6Y5 P91884 A0A034VAF2 A0A1I8P866 W5JC17 A0A182Y7I3 A0A336MJC3 A0A336L8M5 A0A1L8DZG2 A0A3B0KKV3 A0A2M4CIZ1 A0A1W4XAK5 A0A1W4X9D2 Q29FR6 A0A2M4CHN3 A0A2M4CHN1 A0A2M4CHX9 A0A2M3Z590 W5JM65 A0A182PQ49 A0A182RV39 J3JWU4 A0A084VFN0 Q86GB3 A0A2M4AB12 A0A2M4A9Z4 Q86GB2 A0A0N8P0L5 Q8MZM6 A0A0R3P666 A0A2M4AAC2 A0A2M4BJZ2 A0A182UQM2 A0A067RB75 A0A182IA68 A0A182FJY4 A0A2M4BK59 A0A1W4VX70 A0A2M4ADP1 A0A182XLQ9 A0A182NVI1 A0A182W4R7 U5EQ42 A0A2M4AD74 A0A182F127 A0A182MCK5 A0A182QER2 A0A182U8H8 A0A2M4BL19 B4Q0D5 A0A182KEH4 A0A2M4ABX6 B3NX17 B3MXU2 A0A0L0BM47 A0A0Q9WU01 A0A2M3ZGC9 A0A2M4AIK6 A0A2M3Z2G9 A0A0K8TUD9 A0A1W4VJ61 P91938-2 A0A1W4VWE3 A0A0R1EHY5 A0A0R1EBL0 A0A1J1IYM2 A0A1B6CAT4 A0A0P4VIW8 A0A0Q5T6X3 D6WDX1 B4NDH3 A0A0Q5TAX5 A0A1B6CT38 A0A0V0G4M4 A0A182J4F6 A0A1B6BXA1 A0A069DV52 A0A1A9WL68 Q17GT5
PDB
2NVK
E-value=0,
Score=1770
Ontologies
GO
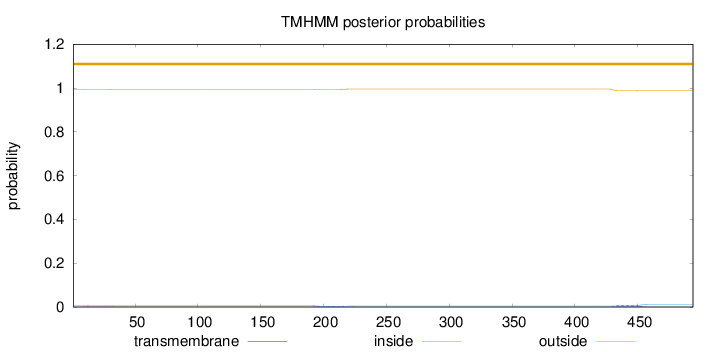
Topology
Subcellular location
Mitochondrion
Cytoplasm
Cytoplasm
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32592
Exp number, first 60 AAs:
0.02269
Total prob of N-in:
0.00714
outside
1 - 494
Population Genetic Test Statistics
Pi
16.29712
Theta
18.769955
Tajima's D
-0.637385
CLR
2.37118
CSRT
0.210589470526474
Interpretation
Uncertain
