Pre Gene Modal
BGIBMGA002886
Annotation
PREDICTED:_C-factor_isoform_X2_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.909
Sequence
CDS
ATGAAGACAGTGCTCATCACAGGAGCCAACAGAGGCCTCGGCCTTGGTATGGTGAAATATCTTACTCACTGCAAAAAGTCGCAGAGAATTATCGCAACTTGCCGAAACGAATCAGAGGAACTAAAACAATTGTCGGAGAAAAATCATAATGTAACAATATTACATTTAGATGTCACAAACTTTTCTGGTTACGATAATTTGACGGCACAGATAAAAAATATCGTCGGAGACCAAGGACTTAATCTGTTAATCAATAATGCCGGTATAACAACCAAATTTACTAAACTGGGGCTGGTGAAGACCGAGCAGCTGATGGACAACCTCACGGTGAACACTGTTGCACCGATCATGCTCACTAAGAGTCTGCTGCCACTATTGAAGCGAGCAGCCGAGCTTTACAGTGGCGAGGCAGTGGGAGTAGAGCGAGCCGCCGTCATCAACATGAGCTCTGTGCTTGGTTCCATAGCGCAAAATGATCAAGGCGGCTTCTACCCCTACAGATGTTCAAAGGCGGCGCTAAACGCGGCAACGAAATCAATGAGCATCGACTTGAAGAAGGAGAGCATCCTCGTTGCTTGTATGCATCCCGGCTGGGTTAAGACCGACATGGGCGGGAAAAACGCTCCCTTAGACGTTGACTCGAGCGTTGCGGGAATTTTCAATACCATAGAGAACTTAGGAGAGGCTGACAGTGGCAAGTTTTTACAATATGACGGTACTGAAATACCTTGGTAA
Protein
MKTVLITGANRGLGLGMVKYLTHCKKSQRIIATCRNESEELKQLSEKNHNVTILHLDVTNFSGYDNLTAQIKNIVGDQGLNLLINNAGITTKFTKLGLVKTEQLMDNLTVNTVAPIMLTKSLLPLLKRAAELYSGEAVGVERAAVINMSSVLGSIAQNDQGGFYPYRCSKAALNAATKSMSIDLKKESILVACMHPGWVKTDMGGKNAPLDVDSSVAGIFNTIENLGEADSGKFLQYDGTEIPW
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J051
S4NTR2
A0A212EHL5
A0A2H1VUE7
I4DJQ8
D2SNU8
+ More
A0A2A4JM24 A0A2P8Z147 A0A026WDR9 A0A3L8DPU0 A0A087ZQ22 A0A2A3EG28 A0A067R941 A0A151IHD3 A0A3Q0J2R3 A0A139WKZ5 E2AHG8 A0A195AW77 T1HQI1 A0A232EV24 A0A023F2E8 J3JYF4 A0A1Q3F9G7 A0A195DMS6 B4JKY1 A0A0M9A310 A0A182GGU9 U4TZY4 A0A1A9WL71 A0A0L7R1C3 B3MXU3 A0A023EL29 B4L1Y4 B4R6H1 A0A1A9Z6D5 A0A0L0BLR9 Q17GT3 A0A0M5J652 A0A2H8TZX2 A0A1W4VIX6 D3TP76 Q9W3H4 B4Q0D7 A0A224XWT4 B3NX16 B4M8A0 E2BEC6 B0WGF8 C4WY93 A0A034VBC9 A0A3B0KYL1 A0A1A9Y480 B4NDH5 A0A1B0FM00 A0A1I8NP09 A0A1A9VLI0 A0A2M3ZBM3 A0A1B0ANI5 A0A154NXY1 A0A2M4AUT8 A0A2M4AVV1 A0A0K8WIX7 E0VUH7 A0A0P4W2Y2 W5JLJ3 B4GU08 A0A182FJY3 A0A1J1IWY7 A0A0A9WCP8 A0A146KPM7 Q29FR5 A0A182PQ48 A0A1I8M0N8 A0A182RV38 A0A2M4BXT0 A0A1L8DCQ3 A0A182IA69 A0A1L8DCV3 A0A182LGZ2 A0A182VE71 A0A182QZ33 A0A084VFN1 A0A182NVI2 A0A182W4R8 A0A182XLR0 W8BI74 A0A182MUW4 Q7PS10 A0A182Y7I4 A0A067RA50 K7IXD6 A0A2J7PD42 A0A1I8NP05 A0A1B0CSU0 A0A2P8Z6R9 A0A158P2K4 A0A2M4CFT4 A0A151WNZ8 T1KIF8 A0A182UG23
A0A2A4JM24 A0A2P8Z147 A0A026WDR9 A0A3L8DPU0 A0A087ZQ22 A0A2A3EG28 A0A067R941 A0A151IHD3 A0A3Q0J2R3 A0A139WKZ5 E2AHG8 A0A195AW77 T1HQI1 A0A232EV24 A0A023F2E8 J3JYF4 A0A1Q3F9G7 A0A195DMS6 B4JKY1 A0A0M9A310 A0A182GGU9 U4TZY4 A0A1A9WL71 A0A0L7R1C3 B3MXU3 A0A023EL29 B4L1Y4 B4R6H1 A0A1A9Z6D5 A0A0L0BLR9 Q17GT3 A0A0M5J652 A0A2H8TZX2 A0A1W4VIX6 D3TP76 Q9W3H4 B4Q0D7 A0A224XWT4 B3NX16 B4M8A0 E2BEC6 B0WGF8 C4WY93 A0A034VBC9 A0A3B0KYL1 A0A1A9Y480 B4NDH5 A0A1B0FM00 A0A1I8NP09 A0A1A9VLI0 A0A2M3ZBM3 A0A1B0ANI5 A0A154NXY1 A0A2M4AUT8 A0A2M4AVV1 A0A0K8WIX7 E0VUH7 A0A0P4W2Y2 W5JLJ3 B4GU08 A0A182FJY3 A0A1J1IWY7 A0A0A9WCP8 A0A146KPM7 Q29FR5 A0A182PQ48 A0A1I8M0N8 A0A182RV38 A0A2M4BXT0 A0A1L8DCQ3 A0A182IA69 A0A1L8DCV3 A0A182LGZ2 A0A182VE71 A0A182QZ33 A0A084VFN1 A0A182NVI2 A0A182W4R8 A0A182XLR0 W8BI74 A0A182MUW4 Q7PS10 A0A182Y7I4 A0A067RA50 K7IXD6 A0A2J7PD42 A0A1I8NP05 A0A1B0CSU0 A0A2P8Z6R9 A0A158P2K4 A0A2M4CFT4 A0A151WNZ8 T1KIF8 A0A182UG23
Pubmed
19121390
23622113
22118469
22651552
26354079
20074338
+ More
29403074 24508170 30249741 24845553 18362917 19820115 20798317 28648823 25474469 22516182 17994087 26483478 23537049 24945155 26108605 17510324 20353571 10731132 12537568 12537572 12537573 12537574 15364585 16110336 17569856 17569867 26109357 26109356 17550304 25348373 20566863 20920257 23761445 25401762 26823975 15632085 25315136 20966253 24438588 24495485 12364791 14747013 17210077 25244985 20075255 21347285
29403074 24508170 30249741 24845553 18362917 19820115 20798317 28648823 25474469 22516182 17994087 26483478 23537049 24945155 26108605 17510324 20353571 10731132 12537568 12537572 12537573 12537574 15364585 16110336 17569856 17569867 26109357 26109356 17550304 25348373 20566863 20920257 23761445 25401762 26823975 15632085 25315136 20966253 24438588 24495485 12364791 14747013 17210077 25244985 20075255 21347285
EMBL
BABH01018954
GAIX01012131
JAA80429.1
AGBW02014864
OWR40978.1
ODYU01004342
+ More
SOQ44112.1 AK401526 KQ459472 BAM18148.1 KPJ00163.1 EZ407232 ACX53789.1 NWSH01001058 PCG72869.1 PYGN01000245 PSN50237.1 KK107293 EZA53164.1 QOIP01000005 RLU22417.1 KZ288254 PBC30715.1 KK852620 KDR20062.1 KQ977614 KYN01380.1 KQ971323 KYB28563.1 GL439539 EFN67123.1 KQ976731 KYM76307.1 ACPB03009184 ACPB03009185 ACPB03009186 NNAY01002061 OXU22204.1 GBBI01003274 JAC15438.1 BT128280 AEE63240.1 GFDL01010878 JAV24167.1 KQ980724 KYN14132.1 CH916370 EDW00234.1 KQ435763 KOX75406.1 JXUM01062535 JXUM01062536 KQ562204 KXJ76421.1 KB631727 ERL85613.1 KQ414668 KOC64591.1 CH902630 EDV38558.1 GAPW01003697 JAC09901.1 CH933810 EDW07705.1 CM000366 EDX17401.1 JRES01001679 KNC21001.1 CH477256 EAT45852.1 CP012528 ALC49094.1 GFXV01007655 MBW19460.1 EZ423228 ADD19504.1 AE014298 AY118557 AAF46353.1 AAM49926.1 AHN59483.1 CM000162 EDX02274.1 GFTR01003985 JAW12441.1 CH954180 EDV46845.1 CH940653 EDW62376.1 GL447768 EFN85971.1 DS231926 EDS26955.1 ABLF02025507 AK343088 BAH72863.1 GAKP01019867 JAC39085.1 OUUW01000018 SPP89208.1 CH964239 EDW82881.1 CCAG010006202 GGFM01005203 MBW25954.1 JXJN01000836 KQ434782 KZC04535.1 GGFK01011222 MBW44543.1 GGFK01011583 MBW44904.1 GDHF01001198 JAI51116.1 DS235786 EEB17033.1 GDRN01076769 JAI62859.1 ADMH02000725 ETN65242.1 CH479190 EDW26028.1 CVRI01000063 CRL04795.1 GBHO01037377 GBRD01002849 JAG06227.1 JAG62972.1 GDHC01020176 JAP98452.1 CH379065 EAL31514.1 GGFJ01008718 MBW57859.1 GFDF01009845 JAV04239.1 APCN01001127 GFDF01009795 JAV04289.1 AXCN02002153 ATLV01012450 KE524793 KFB36775.1 GAMC01005605 JAC00951.1 AXCM01000581 AAAB01008846 EAA06310.3 KK852824 KDR15459.1 AAZX01000012 NEVH01026394 PNF14248.1 AJWK01026583 PYGN01000171 PSN52189.1 ADTU01006914 GGFL01000028 MBW64206.1 KQ982890 KYQ49632.1 CAEY01000112
SOQ44112.1 AK401526 KQ459472 BAM18148.1 KPJ00163.1 EZ407232 ACX53789.1 NWSH01001058 PCG72869.1 PYGN01000245 PSN50237.1 KK107293 EZA53164.1 QOIP01000005 RLU22417.1 KZ288254 PBC30715.1 KK852620 KDR20062.1 KQ977614 KYN01380.1 KQ971323 KYB28563.1 GL439539 EFN67123.1 KQ976731 KYM76307.1 ACPB03009184 ACPB03009185 ACPB03009186 NNAY01002061 OXU22204.1 GBBI01003274 JAC15438.1 BT128280 AEE63240.1 GFDL01010878 JAV24167.1 KQ980724 KYN14132.1 CH916370 EDW00234.1 KQ435763 KOX75406.1 JXUM01062535 JXUM01062536 KQ562204 KXJ76421.1 KB631727 ERL85613.1 KQ414668 KOC64591.1 CH902630 EDV38558.1 GAPW01003697 JAC09901.1 CH933810 EDW07705.1 CM000366 EDX17401.1 JRES01001679 KNC21001.1 CH477256 EAT45852.1 CP012528 ALC49094.1 GFXV01007655 MBW19460.1 EZ423228 ADD19504.1 AE014298 AY118557 AAF46353.1 AAM49926.1 AHN59483.1 CM000162 EDX02274.1 GFTR01003985 JAW12441.1 CH954180 EDV46845.1 CH940653 EDW62376.1 GL447768 EFN85971.1 DS231926 EDS26955.1 ABLF02025507 AK343088 BAH72863.1 GAKP01019867 JAC39085.1 OUUW01000018 SPP89208.1 CH964239 EDW82881.1 CCAG010006202 GGFM01005203 MBW25954.1 JXJN01000836 KQ434782 KZC04535.1 GGFK01011222 MBW44543.1 GGFK01011583 MBW44904.1 GDHF01001198 JAI51116.1 DS235786 EEB17033.1 GDRN01076769 JAI62859.1 ADMH02000725 ETN65242.1 CH479190 EDW26028.1 CVRI01000063 CRL04795.1 GBHO01037377 GBRD01002849 JAG06227.1 JAG62972.1 GDHC01020176 JAP98452.1 CH379065 EAL31514.1 GGFJ01008718 MBW57859.1 GFDF01009845 JAV04239.1 APCN01001127 GFDF01009795 JAV04289.1 AXCN02002153 ATLV01012450 KE524793 KFB36775.1 GAMC01005605 JAC00951.1 AXCM01000581 AAAB01008846 EAA06310.3 KK852824 KDR15459.1 AAZX01000012 NEVH01026394 PNF14248.1 AJWK01026583 PYGN01000171 PSN52189.1 ADTU01006914 GGFL01000028 MBW64206.1 KQ982890 KYQ49632.1 CAEY01000112
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000245037
UP000053097
+ More
UP000279307 UP000005203 UP000242457 UP000027135 UP000078542 UP000079169 UP000007266 UP000000311 UP000078540 UP000015103 UP000215335 UP000078492 UP000001070 UP000053105 UP000069940 UP000249989 UP000030742 UP000091820 UP000053825 UP000007801 UP000009192 UP000000304 UP000092445 UP000037069 UP000008820 UP000092553 UP000192221 UP000000803 UP000002282 UP000008711 UP000008792 UP000008237 UP000002320 UP000007819 UP000268350 UP000092443 UP000007798 UP000092444 UP000095300 UP000078200 UP000092460 UP000076502 UP000009046 UP000000673 UP000008744 UP000069272 UP000183832 UP000001819 UP000075885 UP000095301 UP000075900 UP000075840 UP000075882 UP000075903 UP000075886 UP000030765 UP000075884 UP000075920 UP000076407 UP000075883 UP000007062 UP000076408 UP000002358 UP000235965 UP000092461 UP000005205 UP000075809 UP000015104 UP000075902
UP000279307 UP000005203 UP000242457 UP000027135 UP000078542 UP000079169 UP000007266 UP000000311 UP000078540 UP000015103 UP000215335 UP000078492 UP000001070 UP000053105 UP000069940 UP000249989 UP000030742 UP000091820 UP000053825 UP000007801 UP000009192 UP000000304 UP000092445 UP000037069 UP000008820 UP000092553 UP000192221 UP000000803 UP000002282 UP000008711 UP000008792 UP000008237 UP000002320 UP000007819 UP000268350 UP000092443 UP000007798 UP000092444 UP000095300 UP000078200 UP000092460 UP000076502 UP000009046 UP000000673 UP000008744 UP000069272 UP000183832 UP000001819 UP000075885 UP000095301 UP000075900 UP000075840 UP000075882 UP000075903 UP000075886 UP000030765 UP000075884 UP000075920 UP000076407 UP000075883 UP000007062 UP000076408 UP000002358 UP000235965 UP000092461 UP000005205 UP000075809 UP000015104 UP000075902
Interpro
ProteinModelPortal
H9J051
S4NTR2
A0A212EHL5
A0A2H1VUE7
I4DJQ8
D2SNU8
+ More
A0A2A4JM24 A0A2P8Z147 A0A026WDR9 A0A3L8DPU0 A0A087ZQ22 A0A2A3EG28 A0A067R941 A0A151IHD3 A0A3Q0J2R3 A0A139WKZ5 E2AHG8 A0A195AW77 T1HQI1 A0A232EV24 A0A023F2E8 J3JYF4 A0A1Q3F9G7 A0A195DMS6 B4JKY1 A0A0M9A310 A0A182GGU9 U4TZY4 A0A1A9WL71 A0A0L7R1C3 B3MXU3 A0A023EL29 B4L1Y4 B4R6H1 A0A1A9Z6D5 A0A0L0BLR9 Q17GT3 A0A0M5J652 A0A2H8TZX2 A0A1W4VIX6 D3TP76 Q9W3H4 B4Q0D7 A0A224XWT4 B3NX16 B4M8A0 E2BEC6 B0WGF8 C4WY93 A0A034VBC9 A0A3B0KYL1 A0A1A9Y480 B4NDH5 A0A1B0FM00 A0A1I8NP09 A0A1A9VLI0 A0A2M3ZBM3 A0A1B0ANI5 A0A154NXY1 A0A2M4AUT8 A0A2M4AVV1 A0A0K8WIX7 E0VUH7 A0A0P4W2Y2 W5JLJ3 B4GU08 A0A182FJY3 A0A1J1IWY7 A0A0A9WCP8 A0A146KPM7 Q29FR5 A0A182PQ48 A0A1I8M0N8 A0A182RV38 A0A2M4BXT0 A0A1L8DCQ3 A0A182IA69 A0A1L8DCV3 A0A182LGZ2 A0A182VE71 A0A182QZ33 A0A084VFN1 A0A182NVI2 A0A182W4R8 A0A182XLR0 W8BI74 A0A182MUW4 Q7PS10 A0A182Y7I4 A0A067RA50 K7IXD6 A0A2J7PD42 A0A1I8NP05 A0A1B0CSU0 A0A2P8Z6R9 A0A158P2K4 A0A2M4CFT4 A0A151WNZ8 T1KIF8 A0A182UG23
A0A2A4JM24 A0A2P8Z147 A0A026WDR9 A0A3L8DPU0 A0A087ZQ22 A0A2A3EG28 A0A067R941 A0A151IHD3 A0A3Q0J2R3 A0A139WKZ5 E2AHG8 A0A195AW77 T1HQI1 A0A232EV24 A0A023F2E8 J3JYF4 A0A1Q3F9G7 A0A195DMS6 B4JKY1 A0A0M9A310 A0A182GGU9 U4TZY4 A0A1A9WL71 A0A0L7R1C3 B3MXU3 A0A023EL29 B4L1Y4 B4R6H1 A0A1A9Z6D5 A0A0L0BLR9 Q17GT3 A0A0M5J652 A0A2H8TZX2 A0A1W4VIX6 D3TP76 Q9W3H4 B4Q0D7 A0A224XWT4 B3NX16 B4M8A0 E2BEC6 B0WGF8 C4WY93 A0A034VBC9 A0A3B0KYL1 A0A1A9Y480 B4NDH5 A0A1B0FM00 A0A1I8NP09 A0A1A9VLI0 A0A2M3ZBM3 A0A1B0ANI5 A0A154NXY1 A0A2M4AUT8 A0A2M4AVV1 A0A0K8WIX7 E0VUH7 A0A0P4W2Y2 W5JLJ3 B4GU08 A0A182FJY3 A0A1J1IWY7 A0A0A9WCP8 A0A146KPM7 Q29FR5 A0A182PQ48 A0A1I8M0N8 A0A182RV38 A0A2M4BXT0 A0A1L8DCQ3 A0A182IA69 A0A1L8DCV3 A0A182LGZ2 A0A182VE71 A0A182QZ33 A0A084VFN1 A0A182NVI2 A0A182W4R8 A0A182XLR0 W8BI74 A0A182MUW4 Q7PS10 A0A182Y7I4 A0A067RA50 K7IXD6 A0A2J7PD42 A0A1I8NP05 A0A1B0CSU0 A0A2P8Z6R9 A0A158P2K4 A0A2M4CFT4 A0A151WNZ8 T1KIF8 A0A182UG23
PDB
1SNY
E-value=2.93313e-68,
Score=654
Ontologies
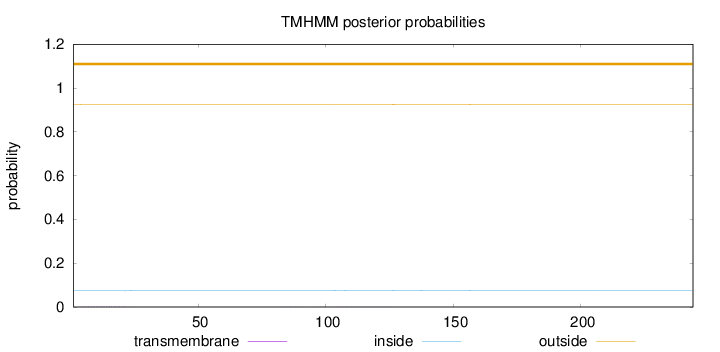
Topology
Length:
244
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04168
Exp number, first 60 AAs:
0.02669
Total prob of N-in:
0.07387
outside
1 - 244
Population Genetic Test Statistics
Pi
15.898722
Theta
15.331118
Tajima's D
-0.883211
CLR
11.784271
CSRT
0.154842257887106
Interpretation
Uncertain
