Gene
KWMTBOMO05983
Pre Gene Modal
BGIBMGA002890
Annotation
gag-pol_polyprotein?_partial_[Lasius_niger]
Location in the cell
Nuclear Reliability : 1.12 PlasmaMembrane Reliability : 1.106
Sequence
CDS
ATGTTTACGTCAGAAAATAATGACGACTATGACTGCTCTTTACTAACATACAAAGAAGCTGTGACTGGCACCGACAGACGAAATTGGGAGAAGGCGATACAATCTGAAATGGAATCATTACAGAAAAATAATACCTGGGTATTAGTAGACAAGTCAGAAGCCAAAGGACACAAAATACTGTCGAACAAATGGGTATTTCGGATTAAAGATGACGGTACGTACAAGGCTAGATTAGTCGTACGTGGATGCGAGCAGAAAGGAAACCTGGATTTCGAAGACATTTATAGCCCAGTAGTAAGTCAATCTGCATTAAAAACTTTATTAGCAATTGCCGCATCTAAATCTTACTACTTCATGACATTTGACATCAAGACAGCTTTCTTATACGGAGAGTTATCAGATGAAGTTTATATGAAATTGCCTGTTGGCTATGAGACTAACTGCAATACTACTGAAGTAGTTTGCCGATTAAAAAAATCTTTGTACGGATTAAAACAAGCTCCATTTACATGGAACAAAACTTTTACCCAAGCGATGTTTGATTTGGGATTCAAGACAATCAAAACAGATCGATGCGTTTTCATAAACGAAGATAAATCAATAATAATCGGACTATTCGTTGATGATGGACTTGTCATAGGAAAAGATAAGAAAAAGATCGAAGCTATTTTGAAGAAACTTGAAACCAAATTTCAAATGAAAAAGAATGAAAATCCAAGTACTTATCTGGGTATGGAAATAAAAAACTCAAAAGAAGGAATAGTTCTTACTCAGAGATCCTACATAATTGACGTACTGGAAAAATACAACATGAAAAATGCCAAACCTTGTGACACGCCCGCAAGTAGCGATTCAAAGTCGATACAGGAAACTGCCCAGGAGAAGGAATATCCCTATAGGGAAGCAGTGGGAAGTTTACTGTACCTGTCTACCAGAACCAGACCAGATATCGCACAAGCGGTCAATCTAGTTAGCAGGAAGGTCGAAAACCCTACTAGAGAAGACGTTACCAAAGTCAAGAAAATATTCAAATATTTAGTGGGAACCAAACATAAAGGCATTTTGTTCAAACGAAACAAACCGATAGAGAGCATCGATGCTTACAGCGACTCGGATTATGCTGGAGACCCATTGACGCGAAGAAGCACTTCTGGTTCCGTGCTAATGATAGCCGATGGACCAATATCATGGGCATCCAAGAGGCAGCCTATAGTAGCGCTATCGTCGACTGAAGCCGAGTACATAGCCGCAGCAGACTGTTGCAAAGAGGCATTGTATCTCAAATCTCTATTACAGGAAACAACCGGTTTGGACATAAAAATAAATCTACATGTTGATAACCAAAGTTCGATTCAACTTGTGAAGTCTGGATCCTTCAACAAAAGCCGCACAGTCTGCGAGTATCGTTGCCGTGAGATGAGTGGTCGCGAGCTGATCGCAGCGTATAAGATATGGCTCTTATTTGGAGCCCTAGCGGAAGTCGTGACGGGCACGGGTTTTGGGAGATGCCCGGCCTACCCGTCTCTACCGAATTTTGACGCACAGAGGATGACCGGCATTTGGTACGAAGTGGAGCGTTCGTTCTACTTGGTGGAGATAGCCGCGAGTTGCACCCGACTTAATGTCACCCTTAACGACAGGGGTTACTTCCAGATCACCGTCGATTCTGTCAACAGATGGACAGGAAGCCAGTCAACTTCATACGGCATCGGCATTCCGTCGTACAATGGGTCGTCAGTGTTTCGATATAAACTAAACAACCGCATGCCTTATTTGATTGGGAGGCTGCTCCCGGGCGCGGGGCAGTACAACATACTCTTCACGGATTACGATCAGTTCGCGCTCGTCTGGTCCTGTAGTAGCGTCAGTCTTGCGCATTCAGATCGAATTTGGGTGTTAGGCCGAAAACAAGAGATCGAAGCGGACGTCCGAGTTCAGATCTACGCCATCATGCAAGAACTGAGACTGGACCCAGACAGGCTGTTCATATCCAAAAACACTAACTGCACCGACTCATTGGAGGCTGAATAA
Protein
MFTSENNDDYDCSLLTYKEAVTGTDRRNWEKAIQSEMESLQKNNTWVLVDKSEAKGHKILSNKWVFRIKDDGTYKARLVVRGCEQKGNLDFEDIYSPVVSQSALKTLLAIAASKSYYFMTFDIKTAFLYGELSDEVYMKLPVGYETNCNTTEVVCRLKKSLYGLKQAPFTWNKTFTQAMFDLGFKTIKTDRCVFINEDKSIIIGLFVDDGLVIGKDKKKIEAILKKLETKFQMKKNENPSTYLGMEIKNSKEGIVLTQRSYIIDVLEKYNMKNAKPCDTPASSDSKSIQETAQEKEYPYREAVGSLLYLSTRTRPDIAQAVNLVSRKVENPTREDVTKVKKIFKYLVGTKHKGILFKRNKPIESIDAYSDSDYAGDPLTRRSTSGSVLMIADGPISWASKRQPIVALSSTEAEYIAAADCCKEALYLKSLLQETTGLDIKINLHVDNQSSIQLVKSGSFNKSRTVCEYRCREMSGRELIAAYKIWLLFGALAEVVTGTGFGRCPAYPSLPNFDAQRMTGIWYEVERSFYLVEIAASCTRLNVTLNDRGYFQITVDSVNRWTGSQSTSYGIGIPSYNGSSVFRYKLNNRMPYLIGRLLPGAGQYNILFTDYDQFALVWSCSSVSLAHSDRIWVLGRKQEIEADVRVQIYAIMQELRLDPDRLFISKNTNCTDSLEAE
Summary
Similarity
Belongs to the terpene synthase family.
Uniprot
A0A0K8TKJ8
A0A139WJX6
A0A2N9GRB2
A0A2N9GCE7
A0A151QND1
A0A2N9I7V5
+ More
A0A1J3DEI6 A0A2N9HZR4 A0A2N9HJ76 A0A2N9GBS9 A0A2N9I6N4 A0A2N9EIR6 A0A2N9FRN5 A0A2N9ELY1 A0A2N9FQJ1 A0A2N9FZ01 A0A2N9FV67 A0A2N9H6B2 A0A2N9I8Q5 A0A2N9HZ75 H9JF18 A0A2N9ETC8 A0A2N9EK90 A0A2N9GGS7 A0A2N9GA80 A0A2N9GBF3 A0A2N9IR42 A0A2N9ID98 A0A2N9EE21 A0A2N9FW82 A0A2N9IGN4 A0A2N9HKJ4 A0A2N9FGL6 A0A2N9J5P5 A0A2N9I0Q5 A0A0J7KRG1
A0A1J3DEI6 A0A2N9HZR4 A0A2N9HJ76 A0A2N9GBS9 A0A2N9I6N4 A0A2N9EIR6 A0A2N9FRN5 A0A2N9ELY1 A0A2N9FQJ1 A0A2N9FZ01 A0A2N9FV67 A0A2N9H6B2 A0A2N9I8Q5 A0A2N9HZ75 H9JF18 A0A2N9ETC8 A0A2N9EK90 A0A2N9GGS7 A0A2N9GA80 A0A2N9GBF3 A0A2N9IR42 A0A2N9ID98 A0A2N9EE21 A0A2N9FW82 A0A2N9IGN4 A0A2N9HKJ4 A0A2N9FGL6 A0A2N9J5P5 A0A2N9I0Q5 A0A0J7KRG1
EMBL
GDAI01002967
JAI14636.1
KQ971338
KYB28117.1
OIVN01002268
SPD02135.1
+ More
OIVN01001729 SPC97049.1 KQ485682 KYP31774.1 OIVN01004910 SPD19931.1 GEVI01015824 JAU16496.1 OIVN01004420 SPD17230.1 OIVN01003897 SPD14257.1 OIVN01001707 SPC96839.1 OIVN01004879 SPD19723.1 OIVN01000116 SPC74628.1 OIVN01001091 SPC89740.1 OIVN01000183 SPC75833.1 OIVN01001052 SPC89229.1 OIVN01001309 SPC92453.1 OIVN01001196 SPC91020.1 OIVN01002904 SPD07378.1 OIVN01005024 SPD20514.1 OIVN01004379 SPD17029.1 BABH01033959 BABH01033960 OIVN01000557 SPC82046.1 OIVN01000147 SPC75222.1 OIVN01001890 SPC98658.1 OIVN01001666 SPC96375.1 OIVN01001669 SPC96464.1 OIVN01006160 SPD26619.1 OIVN01005312 SPD21871.1 OIVN01000036 SPC73022.1 OIVN01001520 SPC94877.1 OIVN01005657 SPD23465.1 OIVN01003569 SPD12194.1 OIVN01000823 SPC86071.1 OIVN01006372 SPD31671.1 OIVN01004867 SPD19656.1 LBMM01003976 KMQ92933.1
OIVN01001729 SPC97049.1 KQ485682 KYP31774.1 OIVN01004910 SPD19931.1 GEVI01015824 JAU16496.1 OIVN01004420 SPD17230.1 OIVN01003897 SPD14257.1 OIVN01001707 SPC96839.1 OIVN01004879 SPD19723.1 OIVN01000116 SPC74628.1 OIVN01001091 SPC89740.1 OIVN01000183 SPC75833.1 OIVN01001052 SPC89229.1 OIVN01001309 SPC92453.1 OIVN01001196 SPC91020.1 OIVN01002904 SPD07378.1 OIVN01005024 SPD20514.1 OIVN01004379 SPD17029.1 BABH01033959 BABH01033960 OIVN01000557 SPC82046.1 OIVN01000147 SPC75222.1 OIVN01001890 SPC98658.1 OIVN01001666 SPC96375.1 OIVN01001669 SPC96464.1 OIVN01006160 SPD26619.1 OIVN01005312 SPD21871.1 OIVN01000036 SPC73022.1 OIVN01001520 SPC94877.1 OIVN01005657 SPD23465.1 OIVN01003569 SPD12194.1 OIVN01000823 SPC86071.1 OIVN01006372 SPD31671.1 OIVN01004867 SPD19656.1 LBMM01003976 KMQ92933.1
Proteomes
PRIDE
Pfam
Interpro
IPR013103
RVT_2
+ More
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR025724 GAG-pre-integrase_dom
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR034741 Terpene_cyclase-like_1_C
IPR036965 Terpene_synth_N_sf
IPR001906 Terpene_synth_N
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR025724 GAG-pre-integrase_dom
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR034741 Terpene_cyclase-like_1_C
IPR036965 Terpene_synth_N_sf
IPR001906 Terpene_synth_N
Gene 3D
ProteinModelPortal
A0A0K8TKJ8
A0A139WJX6
A0A2N9GRB2
A0A2N9GCE7
A0A151QND1
A0A2N9I7V5
+ More
A0A1J3DEI6 A0A2N9HZR4 A0A2N9HJ76 A0A2N9GBS9 A0A2N9I6N4 A0A2N9EIR6 A0A2N9FRN5 A0A2N9ELY1 A0A2N9FQJ1 A0A2N9FZ01 A0A2N9FV67 A0A2N9H6B2 A0A2N9I8Q5 A0A2N9HZ75 H9JF18 A0A2N9ETC8 A0A2N9EK90 A0A2N9GGS7 A0A2N9GA80 A0A2N9GBF3 A0A2N9IR42 A0A2N9ID98 A0A2N9EE21 A0A2N9FW82 A0A2N9IGN4 A0A2N9HKJ4 A0A2N9FGL6 A0A2N9J5P5 A0A2N9I0Q5 A0A0J7KRG1
A0A1J3DEI6 A0A2N9HZR4 A0A2N9HJ76 A0A2N9GBS9 A0A2N9I6N4 A0A2N9EIR6 A0A2N9FRN5 A0A2N9ELY1 A0A2N9FQJ1 A0A2N9FZ01 A0A2N9FV67 A0A2N9H6B2 A0A2N9I8Q5 A0A2N9HZ75 H9JF18 A0A2N9ETC8 A0A2N9EK90 A0A2N9GGS7 A0A2N9GA80 A0A2N9GBF3 A0A2N9IR42 A0A2N9ID98 A0A2N9EE21 A0A2N9FW82 A0A2N9IGN4 A0A2N9HKJ4 A0A2N9FGL6 A0A2N9J5P5 A0A2N9I0Q5 A0A0J7KRG1
PDB
2HZR
E-value=3.05069e-11,
Score=167
Ontologies
KEGG
GO
PANTHER
Topology
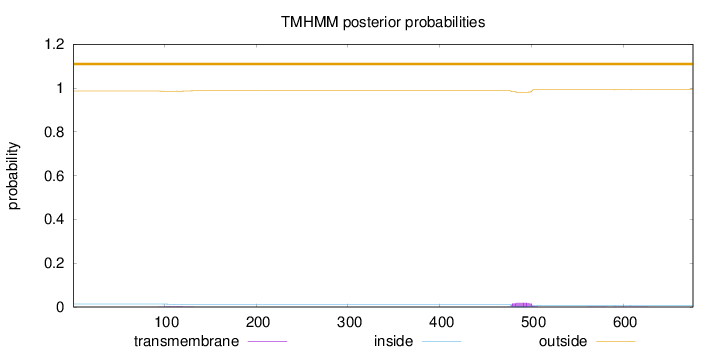
Length:
676
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50238
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01399
outside
1 - 676
Population Genetic Test Statistics
Pi
22.272452
Theta
24.41466
Tajima's D
-0.857142
CLR
1.619475
CSRT
0.165291735413229
Interpretation
Uncertain
