Gene
KWMTBOMO05982
Pre Gene Modal
BGIBMGA002812
Annotation
PREDICTED:_josephin-2_[Amyelois_transitella]
Full name
Autophagy protein 5
Location in the cell
Nuclear Reliability : 2.23
Sequence
CDS
ATGAGTGCGAAACCGACTATTTATCATGAAAAACAAATAAAAGAATTGTGCGCCTTACACGCGCTCAATAATTTATTCCAGACTCGAGGAACATTTTCTAAATCGGAACTAGATGCAATATGCAGTCGCCTGTCTCCAAATGTTTGGATAAATCCTCACAGATCACTTCTTGGCCTTGGAAACTATGATATAAATGTGATAATGGCAGCGTTACAGAAGAAGGGCTGTGAAGCCATCTGGTTTGATAAAAGAAAAGATCCCGGTTGCCTAGATCTTGCCAATATTTGTGGATTCATATTAAATGTGCCTTCGGATTACAAGCTTGGTTTTGTTATGCTGCCATTACGTCGTCGGCATTGGATCACAATTAGACAGATACAGGGAAACTTTTATAATCTTGATTCGAAGTTGGACTCACCTCAGTTAATAGGAAGGAGTTGTGACCTTATCGCGTATCTAAAGGAACAACTAGACAGCAAAGAGAAAGAACTATTTATTGTGGTGAGTAAAGAAGTCGAAGAGAAACAGCTTTGGATGCACCAGTATGTTTTGCCAACACCGGAGCCAAACTATTGCCCTAAACTACCTCTGAACGGTGAAAGTCCAAGCGAAAGCATATCGTCCAACGGCCAATATGATTATAATAATAGAGACGCCGAGTGTCTCAAACTAAATGATGTCCAAAATCATATCCTCAGCACAGAGAATAACTGCTAA
Protein
MSAKPTIYHEKQIKELCALHALNNLFQTRGTFSKSELDAICSRLSPNVWINPHRSLLGLGNYDINVIMAALQKKGCEAIWFDKRKDPGCLDLANICGFILNVPSDYKLGFVMLPLRRRHWITIRQIQGNFYNLDSKLDSPQLIGRSCDLIAYLKEQLDSKEKELFIVVSKEVEEKQLWMHQYVLPTPEPNYCPKLPLNGESPSESISSNGQYDYNNRDAECLKLNDVQNHILSTENNC
Summary
Description
Involved in autophagic vesicle formation.
Subunit
Conjugated with ATG12.
Similarity
Belongs to the ATG5 family.
Uniprot
H9IZX7
A0A3S2P8I0
A0A2H1VR71
A0A2A4J3A8
A0A194QRM0
A0A212FCE5
+ More
A0A194Q3P2 A0A2J7QBI4 A0A224XXG2 A0A069DQ61 A0A023FCU3 A0A0A9W5R0 A0A1Y1N786 R4WNS4 A0A0P4VHG7 T1I1M6 A0A0L7RHM5 A0A170ZT76 E2CA58 A0A151X737 E1ZWU6 A0A026W2D5 A0A067R0N6 F4WWP6 A0A195BK08 A0A195FIC0 A0A158NJ18 A0A195D4W5 A0A1B6IWA0 A0A154P2T5 K7ITM7 A0A0N0BHA5 A0A232EUT6 A0A087ZQ33 A0A2A3E046 V9IKG7 A0A151JMU1 A0A1B6FMZ9 A0A1B6LRM8 A0A0C9RUK9 A0A1B6DIB6 D6X2A7 E9IWK8 A0A0K8TN32 A0A310SQZ0 A0A0T6B351 A0A2M4C102 W5JVH7 T1J928 A0A2M4C0Y4 A0A182WK13 A0A182YC27 A0A182RKW8 A0A182QMS9 A0A2M3ZCG1 A0A2M3ZJH0 A0A182SWI8 A0A1I8NGZ4 A0A182P8U0 A0A182K5M8 A0A023EIQ0 A0A182LBP7 A0A182VKI2 A0A182HNW8 A0A182TQ01 Q7Q9I0 A0A1W4WWH9 A0A182NQ70 A0A1S3K5G6 B4KLR8 W8BCZ7 B4LPH5 A0A0P4WEN3 A0A1Q3F2Q6 B0WMV4 A0A182XEU1 A0A0K8UUB7 A0A0L0C2Z0 A0A2T7PVK7 A0A182MBI7 J3JW92 B7P7H9 A0A1A9V235 A0A1A9X4R1 A0A023G2N4 A0A147BHG7 A0A131XSP0 A0A2P2I8D3 E0VTF4 A0A1B0A3P3 A0A0M4EB42 A0A2R5LKR3 A0A1B0GC96
A0A194Q3P2 A0A2J7QBI4 A0A224XXG2 A0A069DQ61 A0A023FCU3 A0A0A9W5R0 A0A1Y1N786 R4WNS4 A0A0P4VHG7 T1I1M6 A0A0L7RHM5 A0A170ZT76 E2CA58 A0A151X737 E1ZWU6 A0A026W2D5 A0A067R0N6 F4WWP6 A0A195BK08 A0A195FIC0 A0A158NJ18 A0A195D4W5 A0A1B6IWA0 A0A154P2T5 K7ITM7 A0A0N0BHA5 A0A232EUT6 A0A087ZQ33 A0A2A3E046 V9IKG7 A0A151JMU1 A0A1B6FMZ9 A0A1B6LRM8 A0A0C9RUK9 A0A1B6DIB6 D6X2A7 E9IWK8 A0A0K8TN32 A0A310SQZ0 A0A0T6B351 A0A2M4C102 W5JVH7 T1J928 A0A2M4C0Y4 A0A182WK13 A0A182YC27 A0A182RKW8 A0A182QMS9 A0A2M3ZCG1 A0A2M3ZJH0 A0A182SWI8 A0A1I8NGZ4 A0A182P8U0 A0A182K5M8 A0A023EIQ0 A0A182LBP7 A0A182VKI2 A0A182HNW8 A0A182TQ01 Q7Q9I0 A0A1W4WWH9 A0A182NQ70 A0A1S3K5G6 B4KLR8 W8BCZ7 B4LPH5 A0A0P4WEN3 A0A1Q3F2Q6 B0WMV4 A0A182XEU1 A0A0K8UUB7 A0A0L0C2Z0 A0A2T7PVK7 A0A182MBI7 J3JW92 B7P7H9 A0A1A9V235 A0A1A9X4R1 A0A023G2N4 A0A147BHG7 A0A131XSP0 A0A2P2I8D3 E0VTF4 A0A1B0A3P3 A0A0M4EB42 A0A2R5LKR3 A0A1B0GC96
Pubmed
19121390
26354079
22118469
26334808
25474469
25401762
+ More
26823975 28004739 23691247 27129103 20798317 24508170 30249741 24845553 21719571 21347285 20075255 28648823 18362917 19820115 21282665 26369729 20920257 23761445 25244985 25315136 24945155 20966253 12364791 14747013 17210077 17994087 18057021 24495485 26108605 22516182 23537049 29652888 20566863
26823975 28004739 23691247 27129103 20798317 24508170 30249741 24845553 21719571 21347285 20075255 28648823 18362917 19820115 21282665 26369729 20920257 23761445 25244985 25315136 24945155 20966253 12364791 14747013 17210077 17994087 18057021 24495485 26108605 22516182 23537049 29652888 20566863
EMBL
BABH01018973
RSAL01000002
RVE54970.1
ODYU01003953
SOQ43343.1
NWSH01003683
+ More
PCG65930.1 KQ461175 KPJ07979.1 AGBW02009199 OWR51401.1 KQ459472 KPJ00152.1 NEVH01016298 PNF25944.1 GFTR01003204 JAW13222.1 GBGD01003078 JAC85811.1 GBBI01000228 JAC18484.1 GBHO01041766 GDHC01009316 JAG01838.1 JAQ09313.1 GEZM01013070 JAV92780.1 AK417316 BAN20531.1 GDKW01003486 JAI53109.1 ACPB03008059 KQ414592 KOC70236.1 GEMB01001896 JAS01277.1 GL453920 EFN75213.1 KQ982454 KYQ56187.1 GL434905 EFN74343.1 KK107474 QOIP01000002 EZA50158.1 RLU25610.1 KK853027 KDR12294.1 GL888412 EGI61368.1 KQ976453 KYM85508.1 KQ981523 KYN40126.1 ADTU01017351 KQ976885 KYN07469.1 GECU01032844 GECU01016586 JAS74862.1 JAS91120.1 KQ434809 KZC06245.1 KQ435756 KOX75938.1 NNAY01002087 OXU22122.1 KZ288507 PBC25140.1 JR052009 AEY61608.1 KQ978889 KYN27654.1 GECZ01018197 JAS51572.1 GEBQ01031796 GEBQ01024207 GEBQ01013614 GEBQ01010031 JAT08181.1 JAT15770.1 JAT26363.1 JAT29946.1 GBYB01011276 JAG81043.1 GEDC01011896 JAS25402.1 KQ971371 EFA10237.1 GL766581 EFZ15045.1 GDAI01001826 JAI15777.1 KQ761155 OAD58185.1 LJIG01016122 KRT81550.1 GGFJ01009813 MBW58954.1 ADMH02000363 ETN66819.1 JH431968 GGFJ01009814 MBW58955.1 AXCN02001019 GGFM01005441 MBW26192.1 GGFM01007912 MBW28663.1 GAPW01004773 JAC08825.1 APCN01002800 AAAB01008900 EAA09343.3 CH933808 EDW09728.1 KRG04870.1 GAMC01007400 GAMC01007398 JAB99155.1 CH940648 EDW61234.1 GDRN01039947 GDRN01039946 JAI67174.1 GFDL01013200 JAV21845.1 DS232003 EDS31346.1 GDHF01022030 JAI30284.1 JRES01000960 KNC26718.1 PZQS01000001 PVD37465.1 AXCM01012095 APGK01056095 APGK01056096 BT127510 KB741274 KB632130 AEE62472.1 ENN71334.1 ERL88975.1 ABJB010728121 DS652265 EEC02551.1 GBBM01006982 JAC28436.1 GEGO01005230 JAR90174.1 GEFM01005757 JAP70039.1 IACF01004587 LAB70176.1 DS235763 EEB16660.1 CP012524 ALC40677.1 GGLE01005984 MBY10110.1 CCAG010007832
PCG65930.1 KQ461175 KPJ07979.1 AGBW02009199 OWR51401.1 KQ459472 KPJ00152.1 NEVH01016298 PNF25944.1 GFTR01003204 JAW13222.1 GBGD01003078 JAC85811.1 GBBI01000228 JAC18484.1 GBHO01041766 GDHC01009316 JAG01838.1 JAQ09313.1 GEZM01013070 JAV92780.1 AK417316 BAN20531.1 GDKW01003486 JAI53109.1 ACPB03008059 KQ414592 KOC70236.1 GEMB01001896 JAS01277.1 GL453920 EFN75213.1 KQ982454 KYQ56187.1 GL434905 EFN74343.1 KK107474 QOIP01000002 EZA50158.1 RLU25610.1 KK853027 KDR12294.1 GL888412 EGI61368.1 KQ976453 KYM85508.1 KQ981523 KYN40126.1 ADTU01017351 KQ976885 KYN07469.1 GECU01032844 GECU01016586 JAS74862.1 JAS91120.1 KQ434809 KZC06245.1 KQ435756 KOX75938.1 NNAY01002087 OXU22122.1 KZ288507 PBC25140.1 JR052009 AEY61608.1 KQ978889 KYN27654.1 GECZ01018197 JAS51572.1 GEBQ01031796 GEBQ01024207 GEBQ01013614 GEBQ01010031 JAT08181.1 JAT15770.1 JAT26363.1 JAT29946.1 GBYB01011276 JAG81043.1 GEDC01011896 JAS25402.1 KQ971371 EFA10237.1 GL766581 EFZ15045.1 GDAI01001826 JAI15777.1 KQ761155 OAD58185.1 LJIG01016122 KRT81550.1 GGFJ01009813 MBW58954.1 ADMH02000363 ETN66819.1 JH431968 GGFJ01009814 MBW58955.1 AXCN02001019 GGFM01005441 MBW26192.1 GGFM01007912 MBW28663.1 GAPW01004773 JAC08825.1 APCN01002800 AAAB01008900 EAA09343.3 CH933808 EDW09728.1 KRG04870.1 GAMC01007400 GAMC01007398 JAB99155.1 CH940648 EDW61234.1 GDRN01039947 GDRN01039946 JAI67174.1 GFDL01013200 JAV21845.1 DS232003 EDS31346.1 GDHF01022030 JAI30284.1 JRES01000960 KNC26718.1 PZQS01000001 PVD37465.1 AXCM01012095 APGK01056095 APGK01056096 BT127510 KB741274 KB632130 AEE62472.1 ENN71334.1 ERL88975.1 ABJB010728121 DS652265 EEC02551.1 GBBM01006982 JAC28436.1 GEGO01005230 JAR90174.1 GEFM01005757 JAP70039.1 IACF01004587 LAB70176.1 DS235763 EEB16660.1 CP012524 ALC40677.1 GGLE01005984 MBY10110.1 CCAG010007832
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000235965 UP000015103 UP000053825 UP000008237 UP000075809 UP000000311 UP000053097 UP000279307 UP000027135 UP000007755 UP000078540 UP000078541 UP000005205 UP000078542 UP000076502 UP000002358 UP000053105 UP000215335 UP000005203 UP000242457 UP000078492 UP000007266 UP000000673 UP000075920 UP000076408 UP000075900 UP000075886 UP000075901 UP000095301 UP000075885 UP000075881 UP000075882 UP000075903 UP000075840 UP000075902 UP000007062 UP000192223 UP000075884 UP000085678 UP000009192 UP000008792 UP000002320 UP000076407 UP000037069 UP000245119 UP000075883 UP000019118 UP000030742 UP000001555 UP000078200 UP000091820 UP000009046 UP000092445 UP000092553 UP000092444
UP000235965 UP000015103 UP000053825 UP000008237 UP000075809 UP000000311 UP000053097 UP000279307 UP000027135 UP000007755 UP000078540 UP000078541 UP000005205 UP000078542 UP000076502 UP000002358 UP000053105 UP000215335 UP000005203 UP000242457 UP000078492 UP000007266 UP000000673 UP000075920 UP000076408 UP000075900 UP000075886 UP000075901 UP000095301 UP000075885 UP000075881 UP000075882 UP000075903 UP000075840 UP000075902 UP000007062 UP000192223 UP000075884 UP000085678 UP000009192 UP000008792 UP000002320 UP000076407 UP000037069 UP000245119 UP000075883 UP000019118 UP000030742 UP000001555 UP000078200 UP000091820 UP000009046 UP000092445 UP000092553 UP000092444
PRIDE
ProteinModelPortal
H9IZX7
A0A3S2P8I0
A0A2H1VR71
A0A2A4J3A8
A0A194QRM0
A0A212FCE5
+ More
A0A194Q3P2 A0A2J7QBI4 A0A224XXG2 A0A069DQ61 A0A023FCU3 A0A0A9W5R0 A0A1Y1N786 R4WNS4 A0A0P4VHG7 T1I1M6 A0A0L7RHM5 A0A170ZT76 E2CA58 A0A151X737 E1ZWU6 A0A026W2D5 A0A067R0N6 F4WWP6 A0A195BK08 A0A195FIC0 A0A158NJ18 A0A195D4W5 A0A1B6IWA0 A0A154P2T5 K7ITM7 A0A0N0BHA5 A0A232EUT6 A0A087ZQ33 A0A2A3E046 V9IKG7 A0A151JMU1 A0A1B6FMZ9 A0A1B6LRM8 A0A0C9RUK9 A0A1B6DIB6 D6X2A7 E9IWK8 A0A0K8TN32 A0A310SQZ0 A0A0T6B351 A0A2M4C102 W5JVH7 T1J928 A0A2M4C0Y4 A0A182WK13 A0A182YC27 A0A182RKW8 A0A182QMS9 A0A2M3ZCG1 A0A2M3ZJH0 A0A182SWI8 A0A1I8NGZ4 A0A182P8U0 A0A182K5M8 A0A023EIQ0 A0A182LBP7 A0A182VKI2 A0A182HNW8 A0A182TQ01 Q7Q9I0 A0A1W4WWH9 A0A182NQ70 A0A1S3K5G6 B4KLR8 W8BCZ7 B4LPH5 A0A0P4WEN3 A0A1Q3F2Q6 B0WMV4 A0A182XEU1 A0A0K8UUB7 A0A0L0C2Z0 A0A2T7PVK7 A0A182MBI7 J3JW92 B7P7H9 A0A1A9V235 A0A1A9X4R1 A0A023G2N4 A0A147BHG7 A0A131XSP0 A0A2P2I8D3 E0VTF4 A0A1B0A3P3 A0A0M4EB42 A0A2R5LKR3 A0A1B0GC96
A0A194Q3P2 A0A2J7QBI4 A0A224XXG2 A0A069DQ61 A0A023FCU3 A0A0A9W5R0 A0A1Y1N786 R4WNS4 A0A0P4VHG7 T1I1M6 A0A0L7RHM5 A0A170ZT76 E2CA58 A0A151X737 E1ZWU6 A0A026W2D5 A0A067R0N6 F4WWP6 A0A195BK08 A0A195FIC0 A0A158NJ18 A0A195D4W5 A0A1B6IWA0 A0A154P2T5 K7ITM7 A0A0N0BHA5 A0A232EUT6 A0A087ZQ33 A0A2A3E046 V9IKG7 A0A151JMU1 A0A1B6FMZ9 A0A1B6LRM8 A0A0C9RUK9 A0A1B6DIB6 D6X2A7 E9IWK8 A0A0K8TN32 A0A310SQZ0 A0A0T6B351 A0A2M4C102 W5JVH7 T1J928 A0A2M4C0Y4 A0A182WK13 A0A182YC27 A0A182RKW8 A0A182QMS9 A0A2M3ZCG1 A0A2M3ZJH0 A0A182SWI8 A0A1I8NGZ4 A0A182P8U0 A0A182K5M8 A0A023EIQ0 A0A182LBP7 A0A182VKI2 A0A182HNW8 A0A182TQ01 Q7Q9I0 A0A1W4WWH9 A0A182NQ70 A0A1S3K5G6 B4KLR8 W8BCZ7 B4LPH5 A0A0P4WEN3 A0A1Q3F2Q6 B0WMV4 A0A182XEU1 A0A0K8UUB7 A0A0L0C2Z0 A0A2T7PVK7 A0A182MBI7 J3JW92 B7P7H9 A0A1A9V235 A0A1A9X4R1 A0A023G2N4 A0A147BHG7 A0A131XSP0 A0A2P2I8D3 E0VTF4 A0A1B0A3P3 A0A0M4EB42 A0A2R5LKR3 A0A1B0GC96
PDB
3O65
E-value=9.65268e-09,
Score=141
Ontologies
GO
Topology
Subcellular location
Preautophagosomal structure membrane
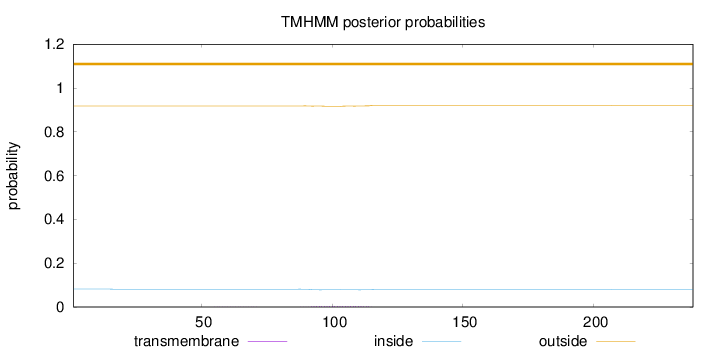
Length:
238
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09695
Exp number, first 60 AAs:
0.00161
Total prob of N-in:
0.08165
outside
1 - 238
Population Genetic Test Statistics
Pi
15.509995
Theta
16.681501
Tajima's D
0.176762
CLR
2.118691
CSRT
0.433228338583071
Interpretation
Uncertain
