Gene
KWMTBOMO05981
Pre Gene Modal
BGIBMGA002891
Annotation
PREDICTED:_fidgetin-like_protein_1_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.5
Sequence
CDS
ATGTCGACAGCTGATGAGAATGATAGACTTTTACTGAATTTTGAAAACATTCAGCAATACAAAAATTCTTTACCAAGTCAATCAAACATTTCCTCTAATGTCATACTCAACAGTGTTTGGAATGAAAACAAGAACATTCTCAATGAAAAAGTTGACGAAATCTTTAAATCCGCTAAAAGCCTTCTCCAAAACTCTGATAGTGAGTTGCATATCAGAACATGGAAATCATCCCTAAGAACTCTAGATCCTGAAATTGGCAAAAATTTTACTATTGAAGAACCCGTTCAAGACTTAGATTTTACAAAAGAAGAACTGGAAAATAAAGCACATAAAAGTAATGTAGCTAAAATTACATTTAAGACTGCAAAAGAACAATTGCTGGCATCTAATCCTGCTGCGAGAAGGACTCTCGGTACTTCTAGGAAAGCTCAAGCCAAGTTTGTGTCTCCCATGATCGGAGCTGTTACTCTAGAAAAACAGGATTCACAAGAAACAGAAATAACTGATGAAAGATTAAAGCATATTGATCCTAAAATGATTGAGTTAATTGAGAGTGAAATTATTGATAAAGGGACACCTGTTAGTTGGGAAGACATTGCCGGGCTGCAGTTCGCCAAGTCTGTGATCCAAGAGGCGGTAGTGTGGCCGCTGCTGAGACCGGACATCTTCACTGGACTGCGGCGGCCCCCCACCGGGATACTGCTGTTCGGGCCACCGGGAACCGGGAAAACATTGATTGGTAAATGCGTGGCGGCGCAGTGCAAGGCTACGTTCTTCAGCATCTCGGCGTCCTCGCTGACGTCCAAGTGGATCGGCGACGGAGAGAAGATGGTGCGCGCGCTGTTCGCAGTGGCGCGGTGCCGCCAACCCGCGGTGATATTCATAGACGAAATAGACTCGTTGCTGACCCAACGCAGCGACTCGGAGCACGAAGCCACCCGGAGAATCAAAACTGAGTTCCTCGTACAATTCGATGGTGCAGCTACAAGTGAAGAAGACAGACTTCTAATAGTTGGCGCGACCAATCGTCCCCAGGAGTTAGACGAAGCTGCCCGCAGGAGACTCGTCAAGCGGCTATACATACCTCTACCAGAAACTGACGCAAGAAAACAAATAATAATGAATTTGCTAAGGAACGAAAACAATGAACTGACGCCGGACCAAATATCGGAAATAGCGGACCTAACTGAAGGGTACTCCGGAGCGGACATGAAGTCTCTGTGCTGCGAGGCTGCCATGGGCCCCGTGCGCGCCGTGCCTCTGTCGCAGATCCTCAACGTCGACCGAGACAAGGTGCGACCAGTTTGTCTCAAAGATTTTACCAGTGCGTTACAAAGGGTCAAGCCGAGTGTATCGCAAGATGATTTGGGACAGTACATCAAATGGAATAAAACTTATGGTCAAGGATTTTAA
Protein
MSTADENDRLLLNFENIQQYKNSLPSQSNISSNVILNSVWNENKNILNEKVDEIFKSAKSLLQNSDSELHIRTWKSSLRTLDPEIGKNFTIEEPVQDLDFTKEELENKAHKSNVAKITFKTAKEQLLASNPAARRTLGTSRKAQAKFVSPMIGAVTLEKQDSQETEITDERLKHIDPKMIELIESEIIDKGTPVSWEDIAGLQFAKSVIQEAVVWPLLRPDIFTGLRRPPTGILLFGPPGTGKTLIGKCVAAQCKATFFSISASSLTSKWIGDGEKMVRALFAVARCRQPAVIFIDEIDSLLTQRSDSEHEATRRIKTEFLVQFDGAATSEEDRLLIVGATNRPQELDEAARRRLVKRLYIPLPETDARKQIIMNLLRNENNELTPDQISEIADLTEGYSGADMKSLCCEAAMGPVRAVPLSQILNVDRDKVRPVCLKDFTSALQRVKPSVSQDDLGQYIKWNKTYGQGF
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9J056
A0A2H1VSZ2
A0A3S2TTP5
S4P8E6
A0A194Q9R3
A0A2A4J240
+ More
A0A212FCE1 Q7PNH8 A0A182TB16 Q16WD0 A0A1S4FLU1 A0A182H821 A0A182W7M9 A0A182XZQ1 A0A182MF44 A0A146L3F9 A0A0K8T9N3 A0A182TGL4 A0A182IZ07 A0A182GXW7 A0A182NTE9 A0A182JVN0 A0A182IED8 A0A182X1F8 A0A1S4GW68 A0A182L2M3 A0A182Q7N0 A0A2J7QHY3 A0A182UM98 A0A0T6B0G4 A0A182RU68 W5JEG7 A0A067RIE0 A0A023F231 D6X4S0 A0A154PEE6 A0A084W7K8 A0A1Y1KPX1 N6TJE9 U5EUR3 A0A0M8ZU70 A0A182F1B9 A0A1S3IGH9 A0A3Q4HRB9 A0A195CJ76 A0A1S3JHD2 A0A3Q2WRF7 A0A3P8QG59 A0A3P9C0X3 A0A3B4F883 E9IKX9 A0A3Q1IEH3 A0A3S1B1R3 A0A3Q1FPF6 A0A151J3K8 A0A151HZH7 A0A158NDM2 A0A3Q0QXT0 A0A3P8RNG7 A0A1B6ERR5 A0A3B4VA14 A0A3B4YGF3 A0A2U9CE67 A0A1B6LYI3 A0A3Q3GBT2 C3YW86 A0A2A3EAD5 K4FUF0 Q4T5A1 I3KZM4 A0A3B4ZGV4 A0A3Q1ALV0 A0A3B4BD04 A0A2D0QL91 A0A0F8C3Z4 A0A088A325 A0A151X906 B0W0A7 B0XF07 A0A3Q3MKU5 A0A369SAM6 A0A3P8VJ85 A0A0N8A7S7 A0A3Q3JF94 A0A3B3YZP3 A0A3B3UW75 A0A195F455 A0A3B3E094 A0A3N0XYR1 A0A1Q3F4J7 A0A3P9NB70 A0A0P5K2T4 B3DGU1 A0A0P5CHN0 E0VN48 A0A3B4DF19
A0A212FCE1 Q7PNH8 A0A182TB16 Q16WD0 A0A1S4FLU1 A0A182H821 A0A182W7M9 A0A182XZQ1 A0A182MF44 A0A146L3F9 A0A0K8T9N3 A0A182TGL4 A0A182IZ07 A0A182GXW7 A0A182NTE9 A0A182JVN0 A0A182IED8 A0A182X1F8 A0A1S4GW68 A0A182L2M3 A0A182Q7N0 A0A2J7QHY3 A0A182UM98 A0A0T6B0G4 A0A182RU68 W5JEG7 A0A067RIE0 A0A023F231 D6X4S0 A0A154PEE6 A0A084W7K8 A0A1Y1KPX1 N6TJE9 U5EUR3 A0A0M8ZU70 A0A182F1B9 A0A1S3IGH9 A0A3Q4HRB9 A0A195CJ76 A0A1S3JHD2 A0A3Q2WRF7 A0A3P8QG59 A0A3P9C0X3 A0A3B4F883 E9IKX9 A0A3Q1IEH3 A0A3S1B1R3 A0A3Q1FPF6 A0A151J3K8 A0A151HZH7 A0A158NDM2 A0A3Q0QXT0 A0A3P8RNG7 A0A1B6ERR5 A0A3B4VA14 A0A3B4YGF3 A0A2U9CE67 A0A1B6LYI3 A0A3Q3GBT2 C3YW86 A0A2A3EAD5 K4FUF0 Q4T5A1 I3KZM4 A0A3B4ZGV4 A0A3Q1ALV0 A0A3B4BD04 A0A2D0QL91 A0A0F8C3Z4 A0A088A325 A0A151X906 B0W0A7 B0XF07 A0A3Q3MKU5 A0A369SAM6 A0A3P8VJ85 A0A0N8A7S7 A0A3Q3JF94 A0A3B3YZP3 A0A3B3UW75 A0A195F455 A0A3B3E094 A0A3N0XYR1 A0A1Q3F4J7 A0A3P9NB70 A0A0P5K2T4 B3DGU1 A0A0P5CHN0 E0VN48 A0A3B4DF19
Pubmed
EMBL
BABH01018974
BABH01018975
ODYU01003953
SOQ43344.1
RSAL01000002
RVE54971.1
+ More
GAIX01005976 JAA86584.1 KQ459472 KPJ00151.1 NWSH01003683 PCG65929.1 AGBW02009199 OWR51402.1 AAAB01008964 EAA12156.5 CH477570 EAT38889.1 JXUM01028692 KQ560829 KXJ80713.1 AXCM01000920 GDHC01015558 JAQ03071.1 GBRD01003564 JAG62257.1 JXUM01020887 JXUM01020888 KQ560584 KXJ81797.1 APCN01005201 AXCN02000707 NEVH01013964 PNF28163.1 LJIG01022445 KRT80574.1 ADMH02001723 ETN61295.1 KK852458 KDR23557.1 GBBI01003723 JAC14989.1 KQ971380 EEZ97654.2 KQ434889 KZC10239.1 ATLV01021261 KE525315 KFB46202.1 GEZM01081273 JAV61725.1 APGK01035525 KB740927 KB631762 ENN77998.1 ERL85920.1 GANO01003755 JAB56116.1 KQ435878 KOX70076.1 KQ977720 KYN00487.1 GL764026 EFZ18713.1 RQTK01001001 RUS72904.1 KQ980275 KYN17018.1 KQ976664 KYM78293.1 ADTU01001047 GECZ01029130 JAS40639.1 CP026257 AWP14360.1 GEBQ01011219 JAT28758.1 GG666560 EEN55415.1 KZ288322 PBC28169.1 JX053327 AFK11555.1 CAAE01009347 CAF91931.1 AERX01006557 KQ042804 KKF10447.1 KQ982402 KYQ56851.1 DS231817 EDS39646.1 DS232886 EDS26355.1 NOWV01000037 RDD43534.1 GDIP01174121 JAJ49281.1 KQ981855 KYN34864.1 RJVU01057857 ROK15766.1 GFDL01012561 JAV22484.1 GDIQ01216108 JAK35617.1 BX890571 BC162519 AAI62519.1 GDIP01174120 JAJ49282.1 DS235332 EEB14814.1
GAIX01005976 JAA86584.1 KQ459472 KPJ00151.1 NWSH01003683 PCG65929.1 AGBW02009199 OWR51402.1 AAAB01008964 EAA12156.5 CH477570 EAT38889.1 JXUM01028692 KQ560829 KXJ80713.1 AXCM01000920 GDHC01015558 JAQ03071.1 GBRD01003564 JAG62257.1 JXUM01020887 JXUM01020888 KQ560584 KXJ81797.1 APCN01005201 AXCN02000707 NEVH01013964 PNF28163.1 LJIG01022445 KRT80574.1 ADMH02001723 ETN61295.1 KK852458 KDR23557.1 GBBI01003723 JAC14989.1 KQ971380 EEZ97654.2 KQ434889 KZC10239.1 ATLV01021261 KE525315 KFB46202.1 GEZM01081273 JAV61725.1 APGK01035525 KB740927 KB631762 ENN77998.1 ERL85920.1 GANO01003755 JAB56116.1 KQ435878 KOX70076.1 KQ977720 KYN00487.1 GL764026 EFZ18713.1 RQTK01001001 RUS72904.1 KQ980275 KYN17018.1 KQ976664 KYM78293.1 ADTU01001047 GECZ01029130 JAS40639.1 CP026257 AWP14360.1 GEBQ01011219 JAT28758.1 GG666560 EEN55415.1 KZ288322 PBC28169.1 JX053327 AFK11555.1 CAAE01009347 CAF91931.1 AERX01006557 KQ042804 KKF10447.1 KQ982402 KYQ56851.1 DS231817 EDS39646.1 DS232886 EDS26355.1 NOWV01000037 RDD43534.1 GDIP01174121 JAJ49281.1 KQ981855 KYN34864.1 RJVU01057857 ROK15766.1 GFDL01012561 JAV22484.1 GDIQ01216108 JAK35617.1 BX890571 BC162519 AAI62519.1 GDIP01174120 JAJ49282.1 DS235332 EEB14814.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000007151
UP000007062
+ More
UP000075901 UP000008820 UP000069940 UP000249989 UP000075920 UP000076408 UP000075883 UP000075902 UP000075880 UP000075884 UP000075881 UP000075840 UP000076407 UP000075882 UP000075886 UP000235965 UP000075903 UP000075900 UP000000673 UP000027135 UP000007266 UP000076502 UP000030765 UP000019118 UP000030742 UP000053105 UP000069272 UP000085678 UP000261580 UP000078542 UP000264840 UP000265100 UP000265160 UP000261460 UP000265040 UP000271974 UP000257200 UP000078492 UP000078540 UP000005205 UP000261340 UP000265080 UP000261420 UP000261360 UP000246464 UP000261660 UP000001554 UP000242457 UP000007303 UP000005207 UP000261400 UP000257160 UP000261520 UP000221080 UP000005203 UP000075809 UP000002320 UP000261640 UP000253843 UP000265120 UP000261600 UP000261480 UP000261500 UP000078541 UP000261560 UP000242638 UP000000437 UP000009046 UP000261440
UP000075901 UP000008820 UP000069940 UP000249989 UP000075920 UP000076408 UP000075883 UP000075902 UP000075880 UP000075884 UP000075881 UP000075840 UP000076407 UP000075882 UP000075886 UP000235965 UP000075903 UP000075900 UP000000673 UP000027135 UP000007266 UP000076502 UP000030765 UP000019118 UP000030742 UP000053105 UP000069272 UP000085678 UP000261580 UP000078542 UP000264840 UP000265100 UP000265160 UP000261460 UP000265040 UP000271974 UP000257200 UP000078492 UP000078540 UP000005205 UP000261340 UP000265080 UP000261420 UP000261360 UP000246464 UP000261660 UP000001554 UP000242457 UP000007303 UP000005207 UP000261400 UP000257160 UP000261520 UP000221080 UP000005203 UP000075809 UP000002320 UP000261640 UP000253843 UP000265120 UP000261600 UP000261480 UP000261500 UP000078541 UP000261560 UP000242638 UP000000437 UP000009046 UP000261440
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J056
A0A2H1VSZ2
A0A3S2TTP5
S4P8E6
A0A194Q9R3
A0A2A4J240
+ More
A0A212FCE1 Q7PNH8 A0A182TB16 Q16WD0 A0A1S4FLU1 A0A182H821 A0A182W7M9 A0A182XZQ1 A0A182MF44 A0A146L3F9 A0A0K8T9N3 A0A182TGL4 A0A182IZ07 A0A182GXW7 A0A182NTE9 A0A182JVN0 A0A182IED8 A0A182X1F8 A0A1S4GW68 A0A182L2M3 A0A182Q7N0 A0A2J7QHY3 A0A182UM98 A0A0T6B0G4 A0A182RU68 W5JEG7 A0A067RIE0 A0A023F231 D6X4S0 A0A154PEE6 A0A084W7K8 A0A1Y1KPX1 N6TJE9 U5EUR3 A0A0M8ZU70 A0A182F1B9 A0A1S3IGH9 A0A3Q4HRB9 A0A195CJ76 A0A1S3JHD2 A0A3Q2WRF7 A0A3P8QG59 A0A3P9C0X3 A0A3B4F883 E9IKX9 A0A3Q1IEH3 A0A3S1B1R3 A0A3Q1FPF6 A0A151J3K8 A0A151HZH7 A0A158NDM2 A0A3Q0QXT0 A0A3P8RNG7 A0A1B6ERR5 A0A3B4VA14 A0A3B4YGF3 A0A2U9CE67 A0A1B6LYI3 A0A3Q3GBT2 C3YW86 A0A2A3EAD5 K4FUF0 Q4T5A1 I3KZM4 A0A3B4ZGV4 A0A3Q1ALV0 A0A3B4BD04 A0A2D0QL91 A0A0F8C3Z4 A0A088A325 A0A151X906 B0W0A7 B0XF07 A0A3Q3MKU5 A0A369SAM6 A0A3P8VJ85 A0A0N8A7S7 A0A3Q3JF94 A0A3B3YZP3 A0A3B3UW75 A0A195F455 A0A3B3E094 A0A3N0XYR1 A0A1Q3F4J7 A0A3P9NB70 A0A0P5K2T4 B3DGU1 A0A0P5CHN0 E0VN48 A0A3B4DF19
A0A212FCE1 Q7PNH8 A0A182TB16 Q16WD0 A0A1S4FLU1 A0A182H821 A0A182W7M9 A0A182XZQ1 A0A182MF44 A0A146L3F9 A0A0K8T9N3 A0A182TGL4 A0A182IZ07 A0A182GXW7 A0A182NTE9 A0A182JVN0 A0A182IED8 A0A182X1F8 A0A1S4GW68 A0A182L2M3 A0A182Q7N0 A0A2J7QHY3 A0A182UM98 A0A0T6B0G4 A0A182RU68 W5JEG7 A0A067RIE0 A0A023F231 D6X4S0 A0A154PEE6 A0A084W7K8 A0A1Y1KPX1 N6TJE9 U5EUR3 A0A0M8ZU70 A0A182F1B9 A0A1S3IGH9 A0A3Q4HRB9 A0A195CJ76 A0A1S3JHD2 A0A3Q2WRF7 A0A3P8QG59 A0A3P9C0X3 A0A3B4F883 E9IKX9 A0A3Q1IEH3 A0A3S1B1R3 A0A3Q1FPF6 A0A151J3K8 A0A151HZH7 A0A158NDM2 A0A3Q0QXT0 A0A3P8RNG7 A0A1B6ERR5 A0A3B4VA14 A0A3B4YGF3 A0A2U9CE67 A0A1B6LYI3 A0A3Q3GBT2 C3YW86 A0A2A3EAD5 K4FUF0 Q4T5A1 I3KZM4 A0A3B4ZGV4 A0A3Q1ALV0 A0A3B4BD04 A0A2D0QL91 A0A0F8C3Z4 A0A088A325 A0A151X906 B0W0A7 B0XF07 A0A3Q3MKU5 A0A369SAM6 A0A3P8VJ85 A0A0N8A7S7 A0A3Q3JF94 A0A3B3YZP3 A0A3B3UW75 A0A195F455 A0A3B3E094 A0A3N0XYR1 A0A1Q3F4J7 A0A3P9NB70 A0A0P5K2T4 B3DGU1 A0A0P5CHN0 E0VN48 A0A3B4DF19
PDB
3D8B
E-value=3.98394e-120,
Score=1105
Ontologies
GO
Topology
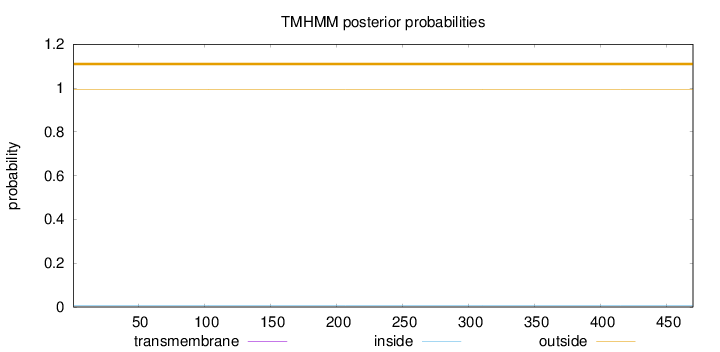
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00264
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00646
outside
1 - 470
Population Genetic Test Statistics
Pi
14.787193
Theta
16.934848
Tajima's D
-0.523827
CLR
10.222698
CSRT
0.24308784560772
Interpretation
Uncertain
