Gene
KWMTBOMO05980
Pre Gene Modal
BGIBMGA002892
Annotation
PREDICTED:_DNA_polymerase_eta_isoform_X2_[Bombyx_mori]
Full name
DNA polymerase eta
Alternative Name
RAD30 homolog A
Xeroderma pigmentosum variant type protein
Xeroderma pigmentosum variant type protein
Location in the cell
Nuclear Reliability : 2.617
Sequence
CDS
ATGGATCAAGACAACAGGGTAGTTGTGTTGATTGATATGGATTGTTTTTATTGTCAAGTTGAAGAGAAACTGAACCCAGAACTGAAAGGAAAACCAATTGCGGTGGTGCAATATAACCCTTGGAAAGGTGGAGGAATAATAGCAGTGAACTATGTTGCGAGGGATTTGGGAGTAACAAGACACATGAGAGGTGATGAGGCCAAAGAGAAATGTCCTAACATTCAGCTGCCTTCAGTTCCTTGTCACAGAGGAAAAGCTGATATATCAAAATATCGAGATGCTGGAAAAGAGGTAGCCAGAGTTTTGCAAAGATTCACAGCTTTGTTAGAAAGAGCTTCAATAGACGAAGCATACCTTGATATTTCAGTGCCAGTGCGCGAAAGACTAAAAACATTAAATATAGGTTCAGTAACTAAAGAAACGTTACCGAATACCTTCGTACTGGGCTACGATTCAATAGAAGAGTTTATGTCTGACGTACTCACGGGTGGTGCAGACTCAATAGATTTTGATTTCGAAGACGCCAAACAGCTGCTGGTAGGTGCTGTCATTGTGAGTGAAATAAGAGCAGCAGTCTATGAAGAAACAGGTTTTCGATGTTCGGCCGGTATAGCTCACAACAAGATCTTGGCGAAGCTGGTGTGCGGCATGAACAAACCCAACAAACAGACAATACTGCCCAAACATTCCATTGATATTCTTTACAGCACGCTGCCAGTGAAAAAGGTTAAACACTTAGGAGGAAAGTTTGGCTCCATGGTATGCACGACACTAAAGATTGCTACAATGGGCGAACTAAAGAAGTTCTCTGTGAAGGAATTGCAGAAGAAGTTTGATGAGAAAAATGGAGCTTGGCTGTACAACGTGGCGCGCGGCATAGACCCGGAGCCGGTTCAGGCCAGGTTTAACCCGAAAAGTATTGGGTGCTGCAAAAATTTTAAAGGCAAATCCGCCCTCGTCGATGTGGACGCGCTCAGGAGATGGCTCAAGGAACTCGCCGACGAAATAGAAAATCGGCTCGATAAAGATGCTCTAGAGAACAGCCGGACGCCGAGACAGATGGTCGCCAGCTACTCCGCGCAGCTCCCGGGAGGCGGGGACTCCGCCGCGTCCAGGTCCTACAACTTTCTGCCTGATGATGACTTAAACGGAGACTTCATGTCCAATAAGGCGTTTGAACTTGTCATGGACGGCTCCGAAGGATACAAACCCAAAGATGTTGAAGACAACAGGAAACTGAAAGCGCCCATAGTTTTTTTGGGAATAAGCGTAGGTAAGTTCGAAGATAATTCTGAAACGAAGAAATCAAAGAAGATCAGGGATTACTTCGAAACTGGAGCTTCTAAGGACATTACGAACAAAGACAATAAAGTATCTTCATTAAAGTCCGAGGAGCTGTGTACGACCGCGCCGCTGGGAACTAACCATTGCAAAGCGAAGACGCTAGAATCGAAAGGAAAAAAAACTGCAGACAAAAGTTACATAATAAATAAATTCTTTAAAGAATCCAACATAAAGACGGAAGCCAATATAATTGAAGAACAAACTGCGCCCTCAGCAGAATATAAGGCGAAATGTGACTCCGTACTAGAGACCAGCTTAGACAGACAAGAATCGTTCTTTGCTAAAATTCTAAACGATAAGTTTAAGGAAAATAGAAATGTACTGAATTCAAAGAGTGACACAGTAATCAAACCGTGCACGCCGGTCTGTGACGTCATCGAGCGTGGCGAAGACAGTGACCAGTCGGATTATTCACGATCGACCATTAACGATGAAATCAATAAAAGTATTGCGCTGTTCGATGAAGATCCTCAGGACGTCCAACGTGTGGCCAGGACTAGGGAATTGCTCAAATCTAATGTTGAAGTAGAAATTGGGACAGCCGGCATTGACGTCGCCGGTCAGTCTGTGAGGGGTGAGGAGAACTACAGTTGTCCTGAATGCGGTAAAGCCGTACCCGTCGGAGATCTAGGAACCCACGCCGACTATCATCTCGCTTTGAACCTAAGAGACGAAGAGAGGCAAACGATTCGACTAGAGCGAGATCAGTTTGAAGCGCGAATTCCTGTTGAAGATAAAAAGAAAAAGAAACCTCCGGACGATATTAATAAAACTGAAAATACAATAGCAAATTTTTTCGTCAAAATCAACGAAAACGTACCGACGGAAACGTGTTCCGAGTGTGGGAGAAAAGTGCAGTTAGATAAGTTTTCCGAACATCTGGACTTCCACGAGGCTCAGAGAATTAGTCGAGAGCTCAACAAGAAGGTTACGGTGAAAATAGGAAGTGGCGAGAAAAGAAAAAGAAAGTCGCCCGTTCATAAATCCAAATTGCCTTGTAAATCTATAACATCATTTTTTAAATAA
Protein
MDQDNRVVVLIDMDCFYCQVEEKLNPELKGKPIAVVQYNPWKGGGIIAVNYVARDLGVTRHMRGDEAKEKCPNIQLPSVPCHRGKADISKYRDAGKEVARVLQRFTALLERASIDEAYLDISVPVRERLKTLNIGSVTKETLPNTFVLGYDSIEEFMSDVLTGGADSIDFDFEDAKQLLVGAVIVSEIRAAVYEETGFRCSAGIAHNKILAKLVCGMNKPNKQTILPKHSIDILYSTLPVKKVKHLGGKFGSMVCTTLKIATMGELKKFSVKELQKKFDEKNGAWLYNVARGIDPEPVQARFNPKSIGCCKNFKGKSALVDVDALRRWLKELADEIENRLDKDALENSRTPRQMVASYSAQLPGGGDSAASRSYNFLPDDDLNGDFMSNKAFELVMDGSEGYKPKDVEDNRKLKAPIVFLGISVGKFEDNSETKKSKKIRDYFETGASKDITNKDNKVSSLKSEELCTTAPLGTNHCKAKTLESKGKKTADKSYIINKFFKESNIKTEANIIEEQTAPSAEYKAKCDSVLETSLDRQESFFAKILNDKFKENRNVLNSKSDTVIKPCTPVCDVIERGEDSDQSDYSRSTINDEINKSIALFDEDPQDVQRVARTRELLKSNVEVEIGTAGIDVAGQSVRGEENYSCPECGKAVPVGDLGTHADYHLALNLRDEERQTIRLERDQFEARIPVEDKKKKKPPDDINKTENTIANFFVKINENVPTETCSECGRKVQLDKFSEHLDFHEAQRISRELNKKVTVKIGSGEKRKRKSPVHKSKLPCKSITSFFK
Summary
Description
DNA polymerase specifically involved in the DNA repair by translesion synthesis (TLS) (PubMed:10385124, PubMed:11743006, PubMed:24449906). Due to low processivity on both damaged and normal DNA, cooperates with the heterotetrameric (REV3L, REV7, POLD2 and POLD3) POLZ complex for complete bypass of DNA lesions. Inserts one or 2 nucleotide(s) opposite the lesion, the primer is further extended by the tetrameric POLZ complex. In the case of 1,2-intrastrand d(GpG)-cisplatin cross-link, inserts dCTP opposite the 3' guanine (PubMed:24449906). Particularly important for the repair of UV-induced pyrimidine dimers (PubMed:10385124, PubMed:11743006). Although inserts the correct base, may cause base transitions and transversions depending upon the context. May play a role in hypermutation at immunoglobulin genes (PubMed:11376341, PubMed:14734526). Forms a Schiff base with 5'-deoxyribose phosphate at abasic sites, but does not have any lyase activity, preventing the release of the 5'-deoxyribose phosphate (5'-dRP) residue. This covalent trapping of the enzyme by the 5'-dRP residue inhibits its DNA synthetic activity during base excision repair, thereby avoiding high incidence of mutagenesis (PubMed:14630940). Targets POLI to replication foci (PubMed:12606586).
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Subunit
Interacts with REV1 (By similarity). Interacts with monoubiquitinated PCNA, but not unmodified PCNA (PubMed:15149598). Interacts with POLI; this interaction targets POLI to the replication machinery (PubMed:12606586). Interacts with PALB2 and BRCA2; the interactions are direct and are required to sustain the recruitment of POLH at blocked replication forks and to stimulate POLH-dependent DNA synthesis on D loop substrates (PubMed:24485656).
Similarity
Belongs to the DNA polymerase type-Y family.
Keywords
3D-structure
Alternative splicing
Complete proteome
Direct protein sequencing
Disease mutation
DNA damage
DNA repair
DNA replication
DNA synthesis
DNA-binding
DNA-directed DNA polymerase
Isopeptide bond
Magnesium
Metal-binding
Mutator protein
Nucleotidyltransferase
Nucleus
Polymorphism
Reference proteome
Schiff base
Transferase
Ubl conjugation
Xeroderma pigmentosum
Feature
chain DNA polymerase eta
splice variant In isoform 2.
sequence variant In XPV.
splice variant In isoform 2.
sequence variant In XPV.
Uniprot
H9J057
A0A2H1VR76
A0A2A4J240
A0A212FCF9
A0A194QR59
A0A194Q568
+ More
B0W8N7 A0A1Q3EY70 W5JMQ9 A0A182P2G4 A0A2M4BDF6 A0A2M4BDF0 A0A2M4BDI7 B4LHT0 A0A182VR94 B4L005 B3MAI8 A0A182YGG4 A0A1A9WSK9 A0A336MV37 A0A1A9VN04 A0A1B0BHB4 A0A0A1XKP3 A0A1A9XBH4 B4IAX3 B4QKP6 A0A3B0J3W2 N6UIC9 B4J0Q9 U4UIC0 A0A336MX77 A0A3L8D811 A0A026VSU6 B4N4G7 A0A0M4EMK3 A0A0J7KX63 A0A151IMR1 A0A2K6SNP8 H2QT29 B3KN75 H9FW04 A0A024RD62 Q9Y253 A0A2K5VUR0 F7HZ00 H9ZAA2 A0A2K5Z3P3 F6SCT4
B0W8N7 A0A1Q3EY70 W5JMQ9 A0A182P2G4 A0A2M4BDF6 A0A2M4BDF0 A0A2M4BDI7 B4LHT0 A0A182VR94 B4L005 B3MAI8 A0A182YGG4 A0A1A9WSK9 A0A336MV37 A0A1A9VN04 A0A1B0BHB4 A0A0A1XKP3 A0A1A9XBH4 B4IAX3 B4QKP6 A0A3B0J3W2 N6UIC9 B4J0Q9 U4UIC0 A0A336MX77 A0A3L8D811 A0A026VSU6 B4N4G7 A0A0M4EMK3 A0A0J7KX63 A0A151IMR1 A0A2K6SNP8 H2QT29 B3KN75 H9FW04 A0A024RD62 Q9Y253 A0A2K5VUR0 F7HZ00 H9ZAA2 A0A2K5Z3P3 F6SCT4
EC Number
2.7.7.7
Pubmed
19121390
22118469
26354079
20920257
23761445
17994087
+ More
25244985 25830018 22936249 23537049 30249741 24508170 16136131 14702039 25319552 11181995 10385124 10398605 11032022 14574404 15489334 11743006 11376341 12606586 14630940 12644469 14734526 15149598 20159558 21791603 22801543 24485656 24449906 11121129 11773631 24130121 16959974 25243066 19892987
25244985 25830018 22936249 23537049 30249741 24508170 16136131 14702039 25319552 11181995 10385124 10398605 11032022 14574404 15489334 11743006 11376341 12606586 14630940 12644469 14734526 15149598 20159558 21791603 22801543 24485656 24449906 11121129 11773631 24130121 16959974 25243066 19892987
EMBL
BABH01018975
BABH01018976
ODYU01003953
SOQ43345.1
NWSH01003683
PCG65929.1
+ More
AGBW02009199 OWR51403.1 KQ461175 KPJ07977.1 KQ459472 KPJ00150.1 DS231859 EDS39087.1 GFDL01014815 JAV20230.1 ADMH02001070 ETN64190.1 GGFJ01001934 MBW51075.1 GGFJ01001935 MBW51076.1 GGFJ01001936 MBW51077.1 CH940647 EDW70655.1 CH933809 EDW19040.1 CH902618 EDV41275.1 UFQS01002906 UFQT01002906 SSX14864.1 SSX34252.1 JXJN01014255 GBXI01002842 JAD11450.1 CH480826 EDW44436.1 CM000363 CM002912 EDX11461.1 KMZ01133.1 OUUW01000002 SPP76404.1 APGK01019170 KB740092 ENN81485.1 CH916366 EDV97914.1 KB632310 ERL92108.1 UFQT01002888 SSX34221.1 QOIP01000012 RLU16232.1 KK108267 EZA46802.1 CH964095 EDW79041.1 CP012525 ALC44947.1 LBMM01002225 KMQ95112.1 KQ977026 KYN06239.1 AACZ04022883 GABC01008341 GABF01003009 GABD01003355 GABE01010341 GABE01010340 GABE01010339 GABE01010338 NBAG03000221 JAA02997.1 JAA19136.1 JAA29745.1 JAA34398.1 PNI77460.1 AK023893 BAG51237.1 JU335060 AFE78813.1 CH471081 EAX04211.1 AB024313 AF158185 AB038008 AY388614 AL353602 AL355802 BC015742 AQIA01051787 AQIA01051788 AQIA01051789 GAMT01009043 GAMS01000370 GAMR01009104 GAMP01010142 JAB02818.1 JAB22766.1 JAB24828.1 JAB42613.1 JU476064 AFH32868.1
AGBW02009199 OWR51403.1 KQ461175 KPJ07977.1 KQ459472 KPJ00150.1 DS231859 EDS39087.1 GFDL01014815 JAV20230.1 ADMH02001070 ETN64190.1 GGFJ01001934 MBW51075.1 GGFJ01001935 MBW51076.1 GGFJ01001936 MBW51077.1 CH940647 EDW70655.1 CH933809 EDW19040.1 CH902618 EDV41275.1 UFQS01002906 UFQT01002906 SSX14864.1 SSX34252.1 JXJN01014255 GBXI01002842 JAD11450.1 CH480826 EDW44436.1 CM000363 CM002912 EDX11461.1 KMZ01133.1 OUUW01000002 SPP76404.1 APGK01019170 KB740092 ENN81485.1 CH916366 EDV97914.1 KB632310 ERL92108.1 UFQT01002888 SSX34221.1 QOIP01000012 RLU16232.1 KK108267 EZA46802.1 CH964095 EDW79041.1 CP012525 ALC44947.1 LBMM01002225 KMQ95112.1 KQ977026 KYN06239.1 AACZ04022883 GABC01008341 GABF01003009 GABD01003355 GABE01010341 GABE01010340 GABE01010339 GABE01010338 NBAG03000221 JAA02997.1 JAA19136.1 JAA29745.1 JAA34398.1 PNI77460.1 AK023893 BAG51237.1 JU335060 AFE78813.1 CH471081 EAX04211.1 AB024313 AF158185 AB038008 AY388614 AL353602 AL355802 BC015742 AQIA01051787 AQIA01051788 AQIA01051789 GAMT01009043 GAMS01000370 GAMR01009104 GAMP01010142 JAB02818.1 JAB22766.1 JAB24828.1 JAB42613.1 JU476064 AFH32868.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000002320
+ More
UP000000673 UP000075885 UP000008792 UP000075920 UP000009192 UP000007801 UP000076408 UP000091820 UP000078200 UP000092460 UP000092443 UP000001292 UP000000304 UP000268350 UP000019118 UP000001070 UP000030742 UP000279307 UP000053097 UP000007798 UP000092553 UP000036403 UP000078542 UP000233220 UP000002277 UP000005640 UP000233100 UP000008225 UP000233140 UP000002281
UP000000673 UP000075885 UP000008792 UP000075920 UP000009192 UP000007801 UP000076408 UP000091820 UP000078200 UP000092460 UP000092443 UP000001292 UP000000304 UP000268350 UP000019118 UP000001070 UP000030742 UP000279307 UP000053097 UP000007798 UP000092553 UP000036403 UP000078542 UP000233220 UP000002277 UP000005640 UP000233100 UP000008225 UP000233140 UP000002281
Interpro
Gene 3D
ProteinModelPortal
H9J057
A0A2H1VR76
A0A2A4J240
A0A212FCF9
A0A194QR59
A0A194Q568
+ More
B0W8N7 A0A1Q3EY70 W5JMQ9 A0A182P2G4 A0A2M4BDF6 A0A2M4BDF0 A0A2M4BDI7 B4LHT0 A0A182VR94 B4L005 B3MAI8 A0A182YGG4 A0A1A9WSK9 A0A336MV37 A0A1A9VN04 A0A1B0BHB4 A0A0A1XKP3 A0A1A9XBH4 B4IAX3 B4QKP6 A0A3B0J3W2 N6UIC9 B4J0Q9 U4UIC0 A0A336MX77 A0A3L8D811 A0A026VSU6 B4N4G7 A0A0M4EMK3 A0A0J7KX63 A0A151IMR1 A0A2K6SNP8 H2QT29 B3KN75 H9FW04 A0A024RD62 Q9Y253 A0A2K5VUR0 F7HZ00 H9ZAA2 A0A2K5Z3P3 F6SCT4
B0W8N7 A0A1Q3EY70 W5JMQ9 A0A182P2G4 A0A2M4BDF6 A0A2M4BDF0 A0A2M4BDI7 B4LHT0 A0A182VR94 B4L005 B3MAI8 A0A182YGG4 A0A1A9WSK9 A0A336MV37 A0A1A9VN04 A0A1B0BHB4 A0A0A1XKP3 A0A1A9XBH4 B4IAX3 B4QKP6 A0A3B0J3W2 N6UIC9 B4J0Q9 U4UIC0 A0A336MX77 A0A3L8D811 A0A026VSU6 B4N4G7 A0A0M4EMK3 A0A0J7KX63 A0A151IMR1 A0A2K6SNP8 H2QT29 B3KN75 H9FW04 A0A024RD62 Q9Y253 A0A2K5VUR0 F7HZ00 H9ZAA2 A0A2K5Z3P3 F6SCT4
PDB
6M7V
E-value=1.09267e-97,
Score=913
Ontologies
GO
GO:0006281
GO:0003684
GO:0005524
GO:0000724
GO:0009650
GO:0019985
GO:0003887
GO:0005634
GO:0016021
GO:0071494
GO:0000731
GO:0006290
GO:0005654
GO:0005829
GO:0006301
GO:0035861
GO:0009314
GO:0005657
GO:0042276
GO:0010225
GO:0006260
GO:0006282
GO:0046872
GO:0070987
GO:0006412
GO:0005622
GO:0008270
GO:0006508
GO:0007264
GO:0008026
GO:0016597
Topology
Subcellular location
Nucleus
Accumulates at replication forks after DNA damage (PubMed:12606586). After UV irradiation, recruited to DNA damage sites within 1 hour, to a maximum of about 80%; this recruitment may not be not restricted to cells active in DNA replication (PubMed:22801543). With evidence from 22 publications.
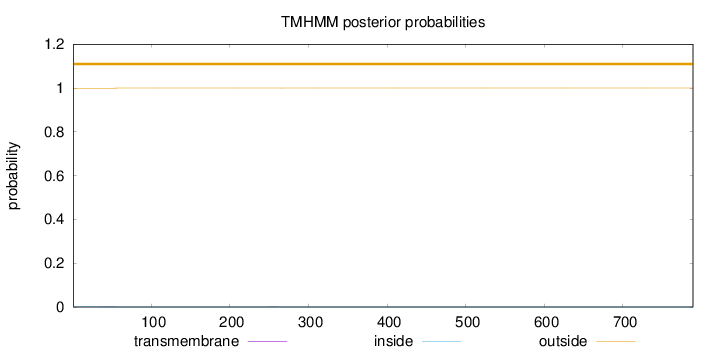
Length:
789
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01265
Exp number, first 60 AAs:
0.01011
Total prob of N-in:
0.00074
outside
1 - 789
Population Genetic Test Statistics
Pi
6.651871
Theta
16.082222
Tajima's D
-1.548667
CLR
12.824101
CSRT
0.0526973651317434
Interpretation
Uncertain
