Pre Gene Modal
BGIBMGA002894
Annotation
PREDICTED:_GTP-binding_protein_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.708
Sequence
CDS
ATGGACAGCGTACCAGAATCTCCGATTAAGGCGATTTCCTTAACTGAGGATGAAGCCTTCGTAGAAAGCGAAACAGATTATTCCAGTATTAGAGGGAAGATGGTGATGGTGAGTCCAAATGCTGATGAGTATGAGTTACTCCGTAAACAGCTATTGGAAAGGCTTGAAGCTGGAAATGGCGAGATCATTTATGATATTGGACATGGAGAAGATGGTCGGGGCTTAACAGAGGATGAATACAATGCGTCCCTAGCAACGCTTCATTCATTGGTGGCAAGTGAGAGAGTGTCATGTGTTGAGCTGAGGAGGTGGGAGTGTGGAAGTGGGCTCACCGGCCAATACCTCCTTAGGACGGAGCTTGACAATAATGACTTTATGGAGATTAGAGTGGCTGTGGTGGGTAACGTGGACGCTGGCAAGTCCACGCTGCTCGGGGTCCTCACGCACGGAGAACTGGACAACGGTCGAGGGTATGCACGACAGAGGCTGTTCCGCCACAAACACGAGATGGAGAGCGGGAGGACCAGCTCCGTGGGGAACGACATATTGGGCTTTGATAATATGGGTAATGTTGTGAATAAGCCAGATCACGGCACTCTTGACTGGGTGAAGATCTGCGAGAAGTCTTCCAAAGTGATAACGTTTATAGACCTAGCTGGACACGAGAGGTACCTCAAAACTACGGTCTTTGGGATGACGGGACACGCTCCCGACTTTGGGATGCTCATGGTTAGTGTTGTAATGCTAAAGATTTTTTTTTTATTCCTTGGATGGGTGAACGAGCTCATAGTCAACCAAGTATTAAGCGGTTACCGGAGCCCATAG
Protein
MDSVPESPIKAISLTEDEAFVESETDYSSIRGKMVMVSPNADEYELLRKQLLERLEAGNGEIIYDIGHGEDGRGLTEDEYNASLATLHSLVASERVSCVELRRWECGSGLTGQYLLRTELDNNDFMEIRVAVVGNVDAGKSTLLGVLTHGELDNGRGYARQRLFRHKHEMESGRTSSVGNDILGFDNMGNVVNKPDHGTLDWVKICEKSSKVITFIDLAGHERYLKTTVFGMTGHAPDFGMLMVSVVMLKIFFLFLGWVNELIVNQVLSGYRSP
Summary
Uniprot
H9J059
A0A2H1WLW7
A0A3S2PLM8
A0A212FCG4
A0A194Q3N7
A0A194QR39
+ More
A0A2J7QBF2 A0A1B6CKT1 B0XK56 T1DPK0 T1DRM8 T1DGA0 A0A232EF70 A0A2J7QBF1 A0A2M3ZAL8 A0A336M7T6 A0A0K8TRU4 A0A1S4EVC2 A0A310SQ58 A0A182SYQ0 Q17PS8 A0A2S2Q811 A0A2P8YJR4 A0A182YC26 A0A151JPI4 A0A154P339 A0A0K8VFQ8 A0A151X1D2 F4W9Y8 E9I9K4 A0A0J7KWN3 A0A158NTC6 A0A182GEP4 A0A151I5T8 A0A023EVQ5 A0A1Q3FWW4 D3TL06 E2B9X0 A0A1A9YD13 A0A1B6IFS0 A0A1B0C274 A0A2M4BFK5 A0A2M4BFK4 A0A151IPM6 A0A1B0GBU0 A0A1A9Z7L5 A0A1A9WQP9 A0A2M4ADA9 A0A1S3DT58 A0A151JU39 A0A1I8Q935 A0A2H8TN13 W5JV36 A0A182FCW7 A0A182RKW9 T1HQL4 A0A084WCP0 W8BM14 V9ICL1 A0A0C9S099 A0A2S2NKW1 A0A1A9VJ01 A0A0A1XH05 A0A1B0EVS3 J9JW71 A0A182QTD8 A0A182JEF1 A0A1B0GM92 A0A087ZQ35 A0A0N0BHA2 A0A023F5K2 A0A0V0G492 A0A0P4VW78 T1PGX4 A0A1I8N4I0 A0A069DZE2 A0A182NQ71 A0A1B6MMP3 A0A182P8U1 A0A1B6G1C6 A0A034WU37 A0A0K8U795 A0A224XL22 A0A182K5M9 A0A182WK12 A0A0A9XRM5 A0A182MGK0 A0A182LBN9 A0A182VJR9 Q7Q9H9 A0A182XEU0 A0A182TN19 A0A182HNW7 A0A2A3E0S5 A0A1J1HK92 E0VCV1 A0A1W4XCJ5 A0A2R7VR23 A0A1Y1JR16
A0A2J7QBF2 A0A1B6CKT1 B0XK56 T1DPK0 T1DRM8 T1DGA0 A0A232EF70 A0A2J7QBF1 A0A2M3ZAL8 A0A336M7T6 A0A0K8TRU4 A0A1S4EVC2 A0A310SQ58 A0A182SYQ0 Q17PS8 A0A2S2Q811 A0A2P8YJR4 A0A182YC26 A0A151JPI4 A0A154P339 A0A0K8VFQ8 A0A151X1D2 F4W9Y8 E9I9K4 A0A0J7KWN3 A0A158NTC6 A0A182GEP4 A0A151I5T8 A0A023EVQ5 A0A1Q3FWW4 D3TL06 E2B9X0 A0A1A9YD13 A0A1B6IFS0 A0A1B0C274 A0A2M4BFK5 A0A2M4BFK4 A0A151IPM6 A0A1B0GBU0 A0A1A9Z7L5 A0A1A9WQP9 A0A2M4ADA9 A0A1S3DT58 A0A151JU39 A0A1I8Q935 A0A2H8TN13 W5JV36 A0A182FCW7 A0A182RKW9 T1HQL4 A0A084WCP0 W8BM14 V9ICL1 A0A0C9S099 A0A2S2NKW1 A0A1A9VJ01 A0A0A1XH05 A0A1B0EVS3 J9JW71 A0A182QTD8 A0A182JEF1 A0A1B0GM92 A0A087ZQ35 A0A0N0BHA2 A0A023F5K2 A0A0V0G492 A0A0P4VW78 T1PGX4 A0A1I8N4I0 A0A069DZE2 A0A182NQ71 A0A1B6MMP3 A0A182P8U1 A0A1B6G1C6 A0A034WU37 A0A0K8U795 A0A224XL22 A0A182K5M9 A0A182WK12 A0A0A9XRM5 A0A182MGK0 A0A182LBN9 A0A182VJR9 Q7Q9H9 A0A182XEU0 A0A182TN19 A0A182HNW7 A0A2A3E0S5 A0A1J1HK92 E0VCV1 A0A1W4XCJ5 A0A2R7VR23 A0A1Y1JR16
Pubmed
19121390
22118469
26354079
24330624
28648823
26369729
+ More
17510324 29403074 25244985 21719571 21282665 21347285 26483478 24945155 20353571 20798317 20920257 23761445 24438588 24495485 25830018 25474469 27129103 25315136 26334808 25348373 25401762 26823975 20966253 12364791 14747013 17210077 20566863 28004739
17510324 29403074 25244985 21719571 21282665 21347285 26483478 24945155 20353571 20798317 20920257 23761445 24438588 24495485 25830018 25474469 27129103 25315136 26334808 25348373 25401762 26823975 20966253 12364791 14747013 17210077 20566863 28004739
EMBL
BABH01018980
ODYU01009515
SOQ54007.1
RSAL01000002
RVE54976.1
AGBW02009199
+ More
OWR51407.1 KQ459472 KPJ00147.1 KQ461175 KPJ07973.1 NEVH01016298 PNF25910.1 GEDC01023231 JAS14067.1 DS233709 EDS31202.1 GAMD01001865 JAA99725.1 GAMD01001859 JAA99731.1 GALA01001756 JAA93096.1 NNAY01005096 OXU17005.1 PNF25911.1 GGFM01004836 MBW25587.1 UFQT01000669 SSX26336.1 GDAI01000521 JAI17082.1 KQ761155 OAD58181.1 CH477189 EAT48763.1 GGMS01004680 MBY73883.1 PYGN01000545 PSN44488.1 KQ978719 KYN29044.1 KQ434809 KZC06242.1 GDHF01014939 JAI37375.1 KQ982585 KYQ54178.1 GL888034 EGI68989.1 GL761846 EFZ22690.1 LBMM01002353 KMQ94937.1 ADTU01025819 ADTU01025820 JXUM01011747 JXUM01011748 KQ560337 KXJ82981.1 KQ976412 KYM90523.1 GAPW01000452 JAC13146.1 GFDL01002945 JAV32100.1 EZ422096 ADD18301.1 GL446605 EFN87557.1 GECU01021936 JAS85770.1 JXJN01024376 GGFJ01002695 MBW51836.1 GGFJ01002694 MBW51835.1 KQ976832 KYN07987.1 CCAG010005890 GGFK01005450 MBW38771.1 KQ981885 KYN33844.1 GFXV01003762 MBW15567.1 ADMH02000363 ETN66820.1 ACPB03000491 ATLV01022798 KE525337 KFB47984.1 GAMC01015857 GAMC01015856 JAB90699.1 JR038760 AEY58397.1 GBYB01013935 GBYB01013936 JAG83702.1 JAG83703.1 GGMR01005158 MBY17777.1 GBXI01004061 GBXI01001561 JAD10231.1 JAD12731.1 AJVK01024086 ABLF02028980 AXCN02001019 AJVK01024087 KQ435756 KOX75942.1 GBBI01002267 JAC16445.1 GECL01003286 JAP02838.1 GDKW01001650 JAI54945.1 KA647138 AFP61767.1 GBGD01000795 JAC88094.1 GEBQ01002796 JAT37181.1 GECZ01013535 JAS56234.1 GAKP01001095 JAC57857.1 GDHF01029787 JAI22527.1 GFTR01007583 JAW08843.1 GBHO01021318 GBHO01021317 GBRD01017283 GDHC01019695 GDHC01007877 JAG22286.1 JAG22287.1 JAG48544.1 JAP98933.1 JAQ10752.1 AXCM01012095 AAAB01008900 EAA09394.4 APCN01002799 KZ288507 PBC25144.1 CVRI01000008 CRK88471.1 DS235065 EEB11207.1 KK854023 PTY09588.1 GEZM01103018 GEZM01103016 JAV51612.1
OWR51407.1 KQ459472 KPJ00147.1 KQ461175 KPJ07973.1 NEVH01016298 PNF25910.1 GEDC01023231 JAS14067.1 DS233709 EDS31202.1 GAMD01001865 JAA99725.1 GAMD01001859 JAA99731.1 GALA01001756 JAA93096.1 NNAY01005096 OXU17005.1 PNF25911.1 GGFM01004836 MBW25587.1 UFQT01000669 SSX26336.1 GDAI01000521 JAI17082.1 KQ761155 OAD58181.1 CH477189 EAT48763.1 GGMS01004680 MBY73883.1 PYGN01000545 PSN44488.1 KQ978719 KYN29044.1 KQ434809 KZC06242.1 GDHF01014939 JAI37375.1 KQ982585 KYQ54178.1 GL888034 EGI68989.1 GL761846 EFZ22690.1 LBMM01002353 KMQ94937.1 ADTU01025819 ADTU01025820 JXUM01011747 JXUM01011748 KQ560337 KXJ82981.1 KQ976412 KYM90523.1 GAPW01000452 JAC13146.1 GFDL01002945 JAV32100.1 EZ422096 ADD18301.1 GL446605 EFN87557.1 GECU01021936 JAS85770.1 JXJN01024376 GGFJ01002695 MBW51836.1 GGFJ01002694 MBW51835.1 KQ976832 KYN07987.1 CCAG010005890 GGFK01005450 MBW38771.1 KQ981885 KYN33844.1 GFXV01003762 MBW15567.1 ADMH02000363 ETN66820.1 ACPB03000491 ATLV01022798 KE525337 KFB47984.1 GAMC01015857 GAMC01015856 JAB90699.1 JR038760 AEY58397.1 GBYB01013935 GBYB01013936 JAG83702.1 JAG83703.1 GGMR01005158 MBY17777.1 GBXI01004061 GBXI01001561 JAD10231.1 JAD12731.1 AJVK01024086 ABLF02028980 AXCN02001019 AJVK01024087 KQ435756 KOX75942.1 GBBI01002267 JAC16445.1 GECL01003286 JAP02838.1 GDKW01001650 JAI54945.1 KA647138 AFP61767.1 GBGD01000795 JAC88094.1 GEBQ01002796 JAT37181.1 GECZ01013535 JAS56234.1 GAKP01001095 JAC57857.1 GDHF01029787 JAI22527.1 GFTR01007583 JAW08843.1 GBHO01021318 GBHO01021317 GBRD01017283 GDHC01019695 GDHC01007877 JAG22286.1 JAG22287.1 JAG48544.1 JAP98933.1 JAQ10752.1 AXCM01012095 AAAB01008900 EAA09394.4 APCN01002799 KZ288507 PBC25144.1 CVRI01000008 CRK88471.1 DS235065 EEB11207.1 KK854023 PTY09588.1 GEZM01103018 GEZM01103016 JAV51612.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000002320 UP000215335 UP000075901 UP000008820 UP000245037 UP000076408 UP000078492 UP000076502 UP000075809 UP000007755 UP000036403 UP000005205 UP000069940 UP000249989 UP000078540 UP000008237 UP000092443 UP000092460 UP000078542 UP000092444 UP000092445 UP000091820 UP000079169 UP000078541 UP000095300 UP000000673 UP000069272 UP000075900 UP000015103 UP000030765 UP000078200 UP000092462 UP000007819 UP000075886 UP000075880 UP000005203 UP000053105 UP000095301 UP000075884 UP000075885 UP000075881 UP000075920 UP000075883 UP000075882 UP000075903 UP000007062 UP000076407 UP000075902 UP000075840 UP000242457 UP000183832 UP000009046 UP000192223
UP000002320 UP000215335 UP000075901 UP000008820 UP000245037 UP000076408 UP000078492 UP000076502 UP000075809 UP000007755 UP000036403 UP000005205 UP000069940 UP000249989 UP000078540 UP000008237 UP000092443 UP000092460 UP000078542 UP000092444 UP000092445 UP000091820 UP000079169 UP000078541 UP000095300 UP000000673 UP000069272 UP000075900 UP000015103 UP000030765 UP000078200 UP000092462 UP000007819 UP000075886 UP000075880 UP000005203 UP000053105 UP000095301 UP000075884 UP000075885 UP000075881 UP000075920 UP000075883 UP000075882 UP000075903 UP000007062 UP000076407 UP000075902 UP000075840 UP000242457 UP000183832 UP000009046 UP000192223
PRIDE
Interpro
ProteinModelPortal
H9J059
A0A2H1WLW7
A0A3S2PLM8
A0A212FCG4
A0A194Q3N7
A0A194QR39
+ More
A0A2J7QBF2 A0A1B6CKT1 B0XK56 T1DPK0 T1DRM8 T1DGA0 A0A232EF70 A0A2J7QBF1 A0A2M3ZAL8 A0A336M7T6 A0A0K8TRU4 A0A1S4EVC2 A0A310SQ58 A0A182SYQ0 Q17PS8 A0A2S2Q811 A0A2P8YJR4 A0A182YC26 A0A151JPI4 A0A154P339 A0A0K8VFQ8 A0A151X1D2 F4W9Y8 E9I9K4 A0A0J7KWN3 A0A158NTC6 A0A182GEP4 A0A151I5T8 A0A023EVQ5 A0A1Q3FWW4 D3TL06 E2B9X0 A0A1A9YD13 A0A1B6IFS0 A0A1B0C274 A0A2M4BFK5 A0A2M4BFK4 A0A151IPM6 A0A1B0GBU0 A0A1A9Z7L5 A0A1A9WQP9 A0A2M4ADA9 A0A1S3DT58 A0A151JU39 A0A1I8Q935 A0A2H8TN13 W5JV36 A0A182FCW7 A0A182RKW9 T1HQL4 A0A084WCP0 W8BM14 V9ICL1 A0A0C9S099 A0A2S2NKW1 A0A1A9VJ01 A0A0A1XH05 A0A1B0EVS3 J9JW71 A0A182QTD8 A0A182JEF1 A0A1B0GM92 A0A087ZQ35 A0A0N0BHA2 A0A023F5K2 A0A0V0G492 A0A0P4VW78 T1PGX4 A0A1I8N4I0 A0A069DZE2 A0A182NQ71 A0A1B6MMP3 A0A182P8U1 A0A1B6G1C6 A0A034WU37 A0A0K8U795 A0A224XL22 A0A182K5M9 A0A182WK12 A0A0A9XRM5 A0A182MGK0 A0A182LBN9 A0A182VJR9 Q7Q9H9 A0A182XEU0 A0A182TN19 A0A182HNW7 A0A2A3E0S5 A0A1J1HK92 E0VCV1 A0A1W4XCJ5 A0A2R7VR23 A0A1Y1JR16
A0A2J7QBF2 A0A1B6CKT1 B0XK56 T1DPK0 T1DRM8 T1DGA0 A0A232EF70 A0A2J7QBF1 A0A2M3ZAL8 A0A336M7T6 A0A0K8TRU4 A0A1S4EVC2 A0A310SQ58 A0A182SYQ0 Q17PS8 A0A2S2Q811 A0A2P8YJR4 A0A182YC26 A0A151JPI4 A0A154P339 A0A0K8VFQ8 A0A151X1D2 F4W9Y8 E9I9K4 A0A0J7KWN3 A0A158NTC6 A0A182GEP4 A0A151I5T8 A0A023EVQ5 A0A1Q3FWW4 D3TL06 E2B9X0 A0A1A9YD13 A0A1B6IFS0 A0A1B0C274 A0A2M4BFK5 A0A2M4BFK4 A0A151IPM6 A0A1B0GBU0 A0A1A9Z7L5 A0A1A9WQP9 A0A2M4ADA9 A0A1S3DT58 A0A151JU39 A0A1I8Q935 A0A2H8TN13 W5JV36 A0A182FCW7 A0A182RKW9 T1HQL4 A0A084WCP0 W8BM14 V9ICL1 A0A0C9S099 A0A2S2NKW1 A0A1A9VJ01 A0A0A1XH05 A0A1B0EVS3 J9JW71 A0A182QTD8 A0A182JEF1 A0A1B0GM92 A0A087ZQ35 A0A0N0BHA2 A0A023F5K2 A0A0V0G492 A0A0P4VW78 T1PGX4 A0A1I8N4I0 A0A069DZE2 A0A182NQ71 A0A1B6MMP3 A0A182P8U1 A0A1B6G1C6 A0A034WU37 A0A0K8U795 A0A224XL22 A0A182K5M9 A0A182WK12 A0A0A9XRM5 A0A182MGK0 A0A182LBN9 A0A182VJR9 Q7Q9H9 A0A182XEU0 A0A182TN19 A0A182HNW7 A0A2A3E0S5 A0A1J1HK92 E0VCV1 A0A1W4XCJ5 A0A2R7VR23 A0A1Y1JR16
PDB
5LZZ
E-value=0.0251646,
Score=86
Ontologies
PANTHER
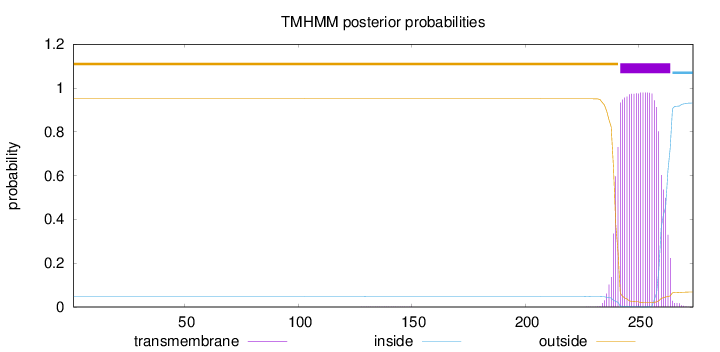
Topology
Length:
274
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.51422
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04895
outside
1 - 241
TMhelix
242 - 264
inside
265 - 274
Population Genetic Test Statistics
Pi
5.119236
Theta
11.964895
Tajima's D
-1.644641
CLR
16.970447
CSRT
0.039798010099495
Interpretation
Uncertain
