Pre Gene Modal
BGIBMGA002895
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_DHX8_[Papilio_xuthus]
Full name
ATP-dependent RNA helicase DHX8
Alternative Name
Peanuts
Location in the cell
Nuclear Reliability : 4.281
Sequence
CDS
ATGGACGAAGTCACTAAATTAGAGCATCTTTCGCTCGTGTCTAAAATATGCACCGAGCTAGACAACCATTTAGGTTTGAACGACAAGGATTTAGCGGAATTCATCATAGATTTGGCTGATAAGAATCCAACATTTGAAACTTTTAAAGTAGCATTGGTTGAAAATGGGGCTGAATTTTCTGATTCCTTCATGACAAATCTACTCCGTATAATACAGCACATGAAACCTGTGAATACATCAACATTGATTAAAAATATTGCCGAAACTGAAAGTGAAAATCCTTTGGCAAAAAAGTTTCCTGGCTTGGCCATACCAAATGATAAACCTCAATCTTTTTCTGATGAAAGTAGTGATGATGAAAAAGACAAAAAGAGAATAACAGCCAAAGACTTATTTACAGAAGAAATTAAACCAAGTTCTACTGATAATGTTGTTGATTTGGCGATGGCTGAATTGGAAGCTTTAGCACCTTCCAATTCTACAACAAATGAACTTCTTATAGGAAAAGAACCCCTAAAAATTGAGAGTAGTAGAAATAGTGATCACAAAGAAAAGAATAAATCAGAAGAATCTAGAGATAAAGAGAGTAGGAGTCGAAATCCTAAAAGAAGCAGAAGTAGGGAACGTAAAAGAAGCAGAAGTACACAGAACACAGATAAATATAAACTAGAAGAATCTCGAGATAGAAAAAGGCATAGTAGAAGTCGAGATCGTAAAAGAAGTAGGAGCCGAGATAGAAAAAGAAGTAGGAGCCGAGATCGTAGAAGAAGTAGGAGTAGAGATTATAAAAGAAGTAGAAGTAGGGAACGTAAAAGAAGTAGAAGTAAAGGTAAGAGAAGTCGTAGCCGGGATAGGAGAAGCAAAAGTCAAGACAGAGACCGTAGAAGAAGATCACACAGTCGACATCGTCACAGATCCAGGTCTAGGGACAGGCGTAGATCTTACTCGAAAAGTAGAAGATCAGTATCAGGTGGCAGAAGATCTCGTTCAAGAAATAGAGAAGAAAAATTTGATTATGGCAAGAAAAGCAGAAAACGCAGCCCTGAAGTAGAATTGAACGATGATCCTGAACCAGGAAAAATATACAGTGGCCGCGTTGCAAACATTGTACCATTCGGCTGTTTTGTTCAAATTGAAGGAGTCAGAAAAAGGTGGGAAGGATTGGTGCATATCTCACAGCTGCGAGCTGAGGGTCGTGTCACCAATGTATCTGATGTTGTCAATAGAGGTGATAAAGTTAAGGTCAAAGTAATCTCAATAACAGGACAGAAAGTTTCTCTCACAATGAAGGATGTTTGTCAAGAGACCGGGAAGGATCTCAATCCAGCATCCCATGCTCACCTGGAAGCAGAACGTGAAGGCCGAAACCCTGATCGCCCTCCACCAGCAGCAACTGTGTTAGCCGGACTCCAAGGTGGAATAGACCCTGATGAGGATTCTAGTAGGAAGAGGGTGACAAGGATATCCAGTCCTGAGAGATGGGAGATCAAACAGATGATTTCATCAGGCGTAATTGATAAGAGTGAATTACCAGATTTTGATGAGGAGACTGGTTTACTTCCAAAAGATGAAGATGGCGAGGCAGACATTGAAATAGAGCTAGTAGAAGAAGAACCTCCTTTCCTCCAAGGTCATGGCAGAGCCCTTCACGATCTTTCTCCAGTAAGGATAGTTAAGAATCCCGATGGCTCCCTTGCACAAGCAGCCATGATGCAGTCCGCATTGGCCAAAGAGAGGAGAGAGCAGAAGATGATCCAACGGGAGCAGGAAATGGACAGCGTACCTACTGGTCTAAATAAAAATTGGATAGATCCGCTACCTGAGTCCGAGGGTCGAACGTTAGCGGCCAACATGCGAGGCGCGGGCGTGGTCCCTCAAGACTTGCCAGAATGGAAGAAACACGTCATCGGTGGAAAAAAATCCTCATTTGGAAAGAAAACGAATTTATCCCTGCTCGAGCAGAGACAGTCGCTGCCCATATACAAGCTTAGAGATGAACTAACTAAGGCCGTGTCCGACAACCAGATCCTGATCGTGATCGGCGAGACGGGGTCGGGCAAGACGACGCAGATAACGCAGTACGTGTGCGAGTGCGGGCTGGCGGCGCGCGGCAAGGTGGCGTGCACGCAGCCGCGCCGCGTGGCCGCCATGTCCGTCGCCAAGCGCGTTGCCGAGGAGTTCGGCTGCCGGCTGGGCCAGGAGGTGGGCTACACCATACGCTTCGAGGACTGCACCAGCCCGGACACTGTCATCAAATACATGACGGACGGCATGCTGCTCCGCGAGTGCCTCATGGACCTGGACCTGAAGGCGTACTCGGTGATCATGCTGGACGAGGCGCACGAGCGCACCATCCACACCGACGTGCTGTTCGGTCTGCTGAAGCAGGCCGTGCAGAAGCGCCCCGAACTCAAGCTCATCGTCACGTCCGCCACGCTCGACGCCGTCAAGTTCTCGCAGTACTTCTTCGAGGCCCCGATCTTCACCATCCCCGGTCGAACCTTCCCCGTCGAAGTCTTGTACACCAAGGAACCGGAGACGGACTATTTGGACGCCTCACTGATCACGGTCATGCAGATCCACCTGAGGGAGCCCCCGGGGGATATCCTGCTGTTCCTCACTGGCCAAGAAGAGATTGACACGGCGTGTGAGATATTGTACGAGAGAATGAAGTCCTTGGGGCCGGACGTGCCGGAGCTGATAATTCTGCCGGTGTACTCTGCGCTGCCGTCCGAGATGCAGACCCGGATCTTCGAGCCGGCGCCCCCCGGCTCCAGGAAGGTGGTGATCGCCACCAACATCGCGGAGACGTCGCTGACCATCGACGGTATATACTACGTGGTGGACCCTGGCTTCGTTAAACAGAAAGTCTACAACTCGAAGACTGGAATGGATTCGTTGGTTGTAACTCCTATATCGCAGGCGGCGGCGAAGCAGCGCGCCGGGCGGGCGGGCCGCACCGGGCCGGGCAAGTGCTACCGGCTGTACACGGAGCGAGCCTACCGGGACGAGATGCTGCCCACGCCCGTGCCCGAGATACAGCGGACCAACCTCGCCACCACCGTATTACAATTGAAAACAATGGGTATAAACGATTTGCTGCATTTCGACTTCATGGACGCGCCGCCGGTCGAGTCGCTCATCATGGCGCTGGAGCAGCTGCACTCGCTGTCGGCGCTCGACTCTGAGGGACTCCTGACGAGGCTGGGACGCAGAATGGCCGAATTTCCACTAGAGCCAAACCTATCGAAGATCCTAATCATGTCGGTGGCCCTGCAGTGCTCGGATGAAATACTCACAATAGTGTCGATGCTGAGCGTCCAAAACGTTTTCTATCGGCCGAAGGACAAGCAGGCGCTTGCCGACCAGAAGAAGGCCAAGTTCAACCAGCCGGAAGGGGACCACCTGACGCTACTGGCCGTGTACAACAGCTGGAGGAATAACAAGTTCTCGAACGCGTGGTGCTACGAAAACTTCGTGCAGATCAGGACGTTGAAGCGGGCCCAGGACGTGAGGAAGCAGCTCCTCGGGATCATGGACAGACACAAATTAGACGTAGTTTCAGCCGGTAAGAATACAGTTCGCATTCAGAAGACGATATGCTCGGGCTTCTTCAGGAACGCGGCCAAGAAGGATCCCCAGGAAGGATATAGGACGCTCGTGGACAGTCAAGTTGTTTATATACATCCTTCCAGTGCACTGTTCAACAGACAACCGGAATGGGTAATCTACCATGAGCTAGTACAGACTACGAAAGAGTACATGAGGGAAGTCACGACCATAGACCCGAAATGGCTGGTTGAGTTTGCTCCCGCCTTCTTTAAATTCTCGGACCCAACAAAACTCTCTAAATTTAAGAAGAACCAGAGACTGGAACCGTTGTACAATAAATATGAGGAGCCGAACGCTTGGAGGATATCGAGAGTCAGACGCAGAAGAAATTAG
Protein
MDEVTKLEHLSLVSKICTELDNHLGLNDKDLAEFIIDLADKNPTFETFKVALVENGAEFSDSFMTNLLRIIQHMKPVNTSTLIKNIAETESENPLAKKFPGLAIPNDKPQSFSDESSDDEKDKKRITAKDLFTEEIKPSSTDNVVDLAMAELEALAPSNSTTNELLIGKEPLKIESSRNSDHKEKNKSEESRDKESRSRNPKRSRSRERKRSRSTQNTDKYKLEESRDRKRHSRSRDRKRSRSRDRKRSRSRDRRRSRSRDYKRSRSRERKRSRSKGKRSRSRDRRSKSQDRDRRRRSHSRHRHRSRSRDRRRSYSKSRRSVSGGRRSRSRNREEKFDYGKKSRKRSPEVELNDDPEPGKIYSGRVANIVPFGCFVQIEGVRKRWEGLVHISQLRAEGRVTNVSDVVNRGDKVKVKVISITGQKVSLTMKDVCQETGKDLNPASHAHLEAEREGRNPDRPPPAATVLAGLQGGIDPDEDSSRKRVTRISSPERWEIKQMISSGVIDKSELPDFDEETGLLPKDEDGEADIEIELVEEEPPFLQGHGRALHDLSPVRIVKNPDGSLAQAAMMQSALAKERREQKMIQREQEMDSVPTGLNKNWIDPLPESEGRTLAANMRGAGVVPQDLPEWKKHVIGGKKSSFGKKTNLSLLEQRQSLPIYKLRDELTKAVSDNQILIVIGETGSGKTTQITQYVCECGLAARGKVACTQPRRVAAMSVAKRVAEEFGCRLGQEVGYTIRFEDCTSPDTVIKYMTDGMLLRECLMDLDLKAYSVIMLDEAHERTIHTDVLFGLLKQAVQKRPELKLIVTSATLDAVKFSQYFFEAPIFTIPGRTFPVEVLYTKEPETDYLDASLITVMQIHLREPPGDILLFLTGQEEIDTACEILYERMKSLGPDVPELIILPVYSALPSEMQTRIFEPAPPGSRKVVIATNIAETSLTIDGIYYVVDPGFVKQKVYNSKTGMDSLVVTPISQAAAKQRAGRAGRTGPGKCYRLYTERAYRDEMLPTPVPEIQRTNLATTVLQLKTMGINDLLHFDFMDAPPVESLIMALEQLHSLSALDSEGLLTRLGRRMAEFPLEPNLSKILIMSVALQCSDEILTIVSMLSVQNVFYRPKDKQALADQKKAKFNQPEGDHLTLLAVYNSWRNNKFSNAWCYENFVQIRTLKRAQDVRKQLLGIMDRHKLDVVSAGKNTVRIQKTICSGFFRNAAKKDPQEGYRTLVDSQVVYIHPSSALFNRQPEWVIYHELVQTTKEYMREVTTIDPKWLVEFAPAFFKFSDPTKLSKFKKNQRLEPLYNKYEEPNAWRISRVRRRRN
Summary
Description
Involved in pre-mRNA splicing as component of the spliceosome (PubMed:18981222, PubMed:24244416). Facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome (By similarity). Before and after egg-chamber formation, required for nurse-cell chromatin dispersal (NCCD) probably by playing a role in spliceosome localization to chromatin/interchromatin spaces (PubMed:24244416).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Identified in the spliceosome C complex.
Similarity
Belongs to the DEAD box helicase family. DEAH subfamily. DDX8/PRP22 sub-subfamily.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Keywords
ATP-binding
Complete proteome
DNA condensation
Helicase
Hydrolase
mRNA processing
mRNA splicing
Nucleotide-binding
Nucleus
Reference proteome
Spliceosome
Feature
chain ATP-dependent RNA helicase DHX8
Uniprot
A0A2H1VKX4
A0A2A4J898
A0A194Q471
A0A1W4XVV8
A0A1Y1JYU8
D6WZK3
+ More
A0A1B6EKN4 B3MD00 A0A2J7QXS3 A0A067QSK0 B4P6M1 B3NRD1 B4KT85 A0A0L0BUI9 A1Z9L3 A0A1W4VCF7 B4LNR3 B4HR09 A0A1I8PWI5 B4MY71 A0A0M9A3B0 A0A1B0FP40 Q292F4 B4JVH2 A0A2A3E6G1 A0A1J1J6C9 A0A1I8MJW9 A0A1B0BUZ3 A0A0M5IY18 A0A182GUX7 A0A088ALV6 B4GDM0 A0A1A9WBZ7 A0A1A9Y2J5 A0A0L7QSX8 A0A154PBE0 A0A1A9VJ30 Q16PT8 A0A182M4X4 A0A0J7NWU6 A0A195C6Y5 A0A182R3J2 A0A3B0J4F9 A0A182YML3 E2AVZ5 A0A182WJR8 A0A151X4R7 A0A026X145 E9IGS5 A0A1Q3F680 E0VM14 A0A158NDX0 A0A084WER8 F4W7U5 A0A195BPP4 A0A182JYP7 A0A182N920 W8C3S0 A0A182PP37 A0A182XLJ4 A0A182V082 E2BY63 A0A182KQN5 A0A182JEE3 A0A182UC60 A0A195D7W1 A0A182I2Q9 A0A182QLE3 A0A195ERA4 Q7PT33 W5J8A4 A0A224XI80 A0A023F5M0 B0WVX3 A0A0K8WDI6 A0A034VE32 A0A2M4A9C4 A0A2M4B9Q7 A0A182FGC7 A0A0A9YMY4 A0A336M1Z5 A0A1B0CYN0 A0A0J9RCJ8 A0A1A9Z7A1 K7J1G4 A0A310SB34 A0A2S2R601 J9K579 T1IZH9 A0A232FI38 T1HWH8 A0A131YM49 A0A224YNR5 A0A131Y0N9 A0A2R5LI29 A0A131XCM6 A0A0P5ZPF8 A0A0P5VN56
A0A1B6EKN4 B3MD00 A0A2J7QXS3 A0A067QSK0 B4P6M1 B3NRD1 B4KT85 A0A0L0BUI9 A1Z9L3 A0A1W4VCF7 B4LNR3 B4HR09 A0A1I8PWI5 B4MY71 A0A0M9A3B0 A0A1B0FP40 Q292F4 B4JVH2 A0A2A3E6G1 A0A1J1J6C9 A0A1I8MJW9 A0A1B0BUZ3 A0A0M5IY18 A0A182GUX7 A0A088ALV6 B4GDM0 A0A1A9WBZ7 A0A1A9Y2J5 A0A0L7QSX8 A0A154PBE0 A0A1A9VJ30 Q16PT8 A0A182M4X4 A0A0J7NWU6 A0A195C6Y5 A0A182R3J2 A0A3B0J4F9 A0A182YML3 E2AVZ5 A0A182WJR8 A0A151X4R7 A0A026X145 E9IGS5 A0A1Q3F680 E0VM14 A0A158NDX0 A0A084WER8 F4W7U5 A0A195BPP4 A0A182JYP7 A0A182N920 W8C3S0 A0A182PP37 A0A182XLJ4 A0A182V082 E2BY63 A0A182KQN5 A0A182JEE3 A0A182UC60 A0A195D7W1 A0A182I2Q9 A0A182QLE3 A0A195ERA4 Q7PT33 W5J8A4 A0A224XI80 A0A023F5M0 B0WVX3 A0A0K8WDI6 A0A034VE32 A0A2M4A9C4 A0A2M4B9Q7 A0A182FGC7 A0A0A9YMY4 A0A336M1Z5 A0A1B0CYN0 A0A0J9RCJ8 A0A1A9Z7A1 K7J1G4 A0A310SB34 A0A2S2R601 J9K579 T1IZH9 A0A232FI38 T1HWH8 A0A131YM49 A0A224YNR5 A0A131Y0N9 A0A2R5LI29 A0A131XCM6 A0A0P5ZPF8 A0A0P5VN56
EC Number
3.6.4.13
Pubmed
26354079
28004739
18362917
19820115
17994087
24845553
+ More
17550304 26108605 10731132 12537572 12537569 18981222 24244416 15632085 25315136 26483478 17510324 25244985 20798317 24508170 30249741 21282665 20566863 21347285 24438588 21719571 24495485 20966253 12364791 14747013 17210077 20920257 23761445 25474469 25348373 25401762 26823975 22936249 20075255 28648823 26830274 28797301 29652888 28049606
17550304 26108605 10731132 12537572 12537569 18981222 24244416 15632085 25315136 26483478 17510324 25244985 20798317 24508170 30249741 21282665 20566863 21347285 24438588 21719571 24495485 20966253 12364791 14747013 17210077 20920257 23761445 25474469 25348373 25401762 26823975 22936249 20075255 28648823 26830274 28797301 29652888 28049606
EMBL
ODYU01003117
SOQ41467.1
NWSH01002488
PCG68191.1
KQ459472
KPJ00144.1
+ More
GEZM01103018 GEZM01103017 GEZM01103016 GEZM01103015 JAV51617.1 KQ971372 EFA09691.1 GECZ01031275 JAS38494.1 CH902619 EDV36315.1 NEVH01009372 PNF33382.1 KK853074 KDR11769.1 CM000158 EDW90973.1 CH954179 EDV56083.1 CH933808 EDW10597.2 JRES01001317 KNC23658.1 AE013599 AY118656 BT044222 AAM50025.1 ACH92287.1 CH940648 EDW60133.1 CH480816 EDW47806.1 CH963894 EDW77060.2 KQ435760 KOX75586.1 CCAG010023115 CM000071 EAL24908.1 CH916375 EDV98440.1 KZ288353 PBC27300.1 CVRI01000069 CRL07030.1 JXJN01020988 CP012524 ALC42456.1 JXUM01018693 JXUM01018694 KQ560519 KXJ82015.1 CH479181 EDW31677.1 KQ414756 KOC61659.1 KQ434868 KZC09161.1 CH477773 EAT36372.1 AXCM01002191 LBMM01001117 KMQ96895.1 KQ978251 KYM95936.1 OUUW01000001 SPP74383.1 GL443213 EFN62417.1 KQ982548 KYQ55280.1 KK107046 QOIP01000004 EZA61721.1 RLU23672.1 GL763054 EFZ20200.1 GFDL01011974 JAV23071.1 DS235286 EEB14420.1 ADTU01013059 ATLV01023246 KE525341 KFB48712.1 GL887888 EGI69615.1 KQ976424 KYM88480.1 GAMC01002647 JAC03909.1 GL451420 EFN79342.1 KQ981153 KYN08975.1 APCN01000178 AXCN02000540 KQ981993 KYN30790.1 AAAB01008807 EAA04624.5 ADMH02002070 ETN59623.1 GFTR01008583 JAW07843.1 GBBI01002253 JAC16459.1 DS232134 EDS35805.1 GDHF01003404 JAI48910.1 GAKP01019149 JAC39803.1 GGFK01004066 MBW37387.1 GGFJ01000601 MBW49742.1 GBHO01010066 GBHO01010065 GBRD01005381 GDHC01020627 GDHC01004880 JAG33538.1 JAG33539.1 JAG60440.1 JAP98001.1 JAQ13749.1 UFQT01000289 SSX22853.1 AJVK01009248 CM002911 KMY93686.1 KQ773953 OAD52333.1 GGMS01016150 MBY85353.1 ABLF02023620 AFFK01020457 NNAY01000193 OXU30118.1 ACPB03016233 GEDV01008935 JAP79622.1 GFPF01006243 MAA17389.1 GEFM01003761 GEGO01006586 JAP72035.1 JAR88818.1 GGLE01005030 MBY09156.1 GEFH01004731 JAP63850.1 GDIP01044533 JAM59182.1 GDIP01097776 JAM05939.1
GEZM01103018 GEZM01103017 GEZM01103016 GEZM01103015 JAV51617.1 KQ971372 EFA09691.1 GECZ01031275 JAS38494.1 CH902619 EDV36315.1 NEVH01009372 PNF33382.1 KK853074 KDR11769.1 CM000158 EDW90973.1 CH954179 EDV56083.1 CH933808 EDW10597.2 JRES01001317 KNC23658.1 AE013599 AY118656 BT044222 AAM50025.1 ACH92287.1 CH940648 EDW60133.1 CH480816 EDW47806.1 CH963894 EDW77060.2 KQ435760 KOX75586.1 CCAG010023115 CM000071 EAL24908.1 CH916375 EDV98440.1 KZ288353 PBC27300.1 CVRI01000069 CRL07030.1 JXJN01020988 CP012524 ALC42456.1 JXUM01018693 JXUM01018694 KQ560519 KXJ82015.1 CH479181 EDW31677.1 KQ414756 KOC61659.1 KQ434868 KZC09161.1 CH477773 EAT36372.1 AXCM01002191 LBMM01001117 KMQ96895.1 KQ978251 KYM95936.1 OUUW01000001 SPP74383.1 GL443213 EFN62417.1 KQ982548 KYQ55280.1 KK107046 QOIP01000004 EZA61721.1 RLU23672.1 GL763054 EFZ20200.1 GFDL01011974 JAV23071.1 DS235286 EEB14420.1 ADTU01013059 ATLV01023246 KE525341 KFB48712.1 GL887888 EGI69615.1 KQ976424 KYM88480.1 GAMC01002647 JAC03909.1 GL451420 EFN79342.1 KQ981153 KYN08975.1 APCN01000178 AXCN02000540 KQ981993 KYN30790.1 AAAB01008807 EAA04624.5 ADMH02002070 ETN59623.1 GFTR01008583 JAW07843.1 GBBI01002253 JAC16459.1 DS232134 EDS35805.1 GDHF01003404 JAI48910.1 GAKP01019149 JAC39803.1 GGFK01004066 MBW37387.1 GGFJ01000601 MBW49742.1 GBHO01010066 GBHO01010065 GBRD01005381 GDHC01020627 GDHC01004880 JAG33538.1 JAG33539.1 JAG60440.1 JAP98001.1 JAQ13749.1 UFQT01000289 SSX22853.1 AJVK01009248 CM002911 KMY93686.1 KQ773953 OAD52333.1 GGMS01016150 MBY85353.1 ABLF02023620 AFFK01020457 NNAY01000193 OXU30118.1 ACPB03016233 GEDV01008935 JAP79622.1 GFPF01006243 MAA17389.1 GEFM01003761 GEGO01006586 JAP72035.1 JAR88818.1 GGLE01005030 MBY09156.1 GEFH01004731 JAP63850.1 GDIP01044533 JAM59182.1 GDIP01097776 JAM05939.1
Proteomes
UP000218220
UP000053268
UP000192223
UP000007266
UP000007801
UP000235965
+ More
UP000027135 UP000002282 UP000008711 UP000009192 UP000037069 UP000000803 UP000192221 UP000008792 UP000001292 UP000095300 UP000007798 UP000053105 UP000092444 UP000001819 UP000001070 UP000242457 UP000183832 UP000095301 UP000092460 UP000092553 UP000069940 UP000249989 UP000005203 UP000008744 UP000091820 UP000092443 UP000053825 UP000076502 UP000078200 UP000008820 UP000075883 UP000036403 UP000078542 UP000075900 UP000268350 UP000076408 UP000000311 UP000075920 UP000075809 UP000053097 UP000279307 UP000009046 UP000005205 UP000030765 UP000007755 UP000078540 UP000075881 UP000075884 UP000075885 UP000076407 UP000075903 UP000008237 UP000075882 UP000075880 UP000075902 UP000078492 UP000075840 UP000075886 UP000078541 UP000007062 UP000000673 UP000002320 UP000069272 UP000092462 UP000092445 UP000002358 UP000007819 UP000215335 UP000015103
UP000027135 UP000002282 UP000008711 UP000009192 UP000037069 UP000000803 UP000192221 UP000008792 UP000001292 UP000095300 UP000007798 UP000053105 UP000092444 UP000001819 UP000001070 UP000242457 UP000183832 UP000095301 UP000092460 UP000092553 UP000069940 UP000249989 UP000005203 UP000008744 UP000091820 UP000092443 UP000053825 UP000076502 UP000078200 UP000008820 UP000075883 UP000036403 UP000078542 UP000075900 UP000268350 UP000076408 UP000000311 UP000075920 UP000075809 UP000053097 UP000279307 UP000009046 UP000005205 UP000030765 UP000007755 UP000078540 UP000075881 UP000075884 UP000075885 UP000076407 UP000075903 UP000008237 UP000075882 UP000075880 UP000075902 UP000078492 UP000075840 UP000075886 UP000078541 UP000007062 UP000000673 UP000002320 UP000069272 UP000092462 UP000092445 UP000002358 UP000007819 UP000215335 UP000015103
PRIDE
Pfam
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR011545 DEAD/DEAH_box_helicase_dom
IPR012340 NA-bd_OB-fold
IPR011709 DUF1605
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR007502 Helicase-assoc_dom
IPR001841 Znf_RING
IPR003029 S1_domain
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR022967 S1_dom
IPR022170 GIDE
IPR042035 DEAH_win-hel_dom
IPR004328 BRO1_dom
IPR038499 BRO1_sf
IPR025304 ALIX_V_dom
IPR001737 KsgA/Erm
IPR029063 SAM-dependent_MTases
IPR011545 DEAD/DEAH_box_helicase_dom
IPR012340 NA-bd_OB-fold
IPR011709 DUF1605
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR007502 Helicase-assoc_dom
IPR001841 Znf_RING
IPR003029 S1_domain
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR022967 S1_dom
IPR022170 GIDE
IPR042035 DEAH_win-hel_dom
IPR004328 BRO1_dom
IPR038499 BRO1_sf
IPR025304 ALIX_V_dom
IPR001737 KsgA/Erm
IPR029063 SAM-dependent_MTases
Gene 3D
CDD
ProteinModelPortal
A0A2H1VKX4
A0A2A4J898
A0A194Q471
A0A1W4XVV8
A0A1Y1JYU8
D6WZK3
+ More
A0A1B6EKN4 B3MD00 A0A2J7QXS3 A0A067QSK0 B4P6M1 B3NRD1 B4KT85 A0A0L0BUI9 A1Z9L3 A0A1W4VCF7 B4LNR3 B4HR09 A0A1I8PWI5 B4MY71 A0A0M9A3B0 A0A1B0FP40 Q292F4 B4JVH2 A0A2A3E6G1 A0A1J1J6C9 A0A1I8MJW9 A0A1B0BUZ3 A0A0M5IY18 A0A182GUX7 A0A088ALV6 B4GDM0 A0A1A9WBZ7 A0A1A9Y2J5 A0A0L7QSX8 A0A154PBE0 A0A1A9VJ30 Q16PT8 A0A182M4X4 A0A0J7NWU6 A0A195C6Y5 A0A182R3J2 A0A3B0J4F9 A0A182YML3 E2AVZ5 A0A182WJR8 A0A151X4R7 A0A026X145 E9IGS5 A0A1Q3F680 E0VM14 A0A158NDX0 A0A084WER8 F4W7U5 A0A195BPP4 A0A182JYP7 A0A182N920 W8C3S0 A0A182PP37 A0A182XLJ4 A0A182V082 E2BY63 A0A182KQN5 A0A182JEE3 A0A182UC60 A0A195D7W1 A0A182I2Q9 A0A182QLE3 A0A195ERA4 Q7PT33 W5J8A4 A0A224XI80 A0A023F5M0 B0WVX3 A0A0K8WDI6 A0A034VE32 A0A2M4A9C4 A0A2M4B9Q7 A0A182FGC7 A0A0A9YMY4 A0A336M1Z5 A0A1B0CYN0 A0A0J9RCJ8 A0A1A9Z7A1 K7J1G4 A0A310SB34 A0A2S2R601 J9K579 T1IZH9 A0A232FI38 T1HWH8 A0A131YM49 A0A224YNR5 A0A131Y0N9 A0A2R5LI29 A0A131XCM6 A0A0P5ZPF8 A0A0P5VN56
A0A1B6EKN4 B3MD00 A0A2J7QXS3 A0A067QSK0 B4P6M1 B3NRD1 B4KT85 A0A0L0BUI9 A1Z9L3 A0A1W4VCF7 B4LNR3 B4HR09 A0A1I8PWI5 B4MY71 A0A0M9A3B0 A0A1B0FP40 Q292F4 B4JVH2 A0A2A3E6G1 A0A1J1J6C9 A0A1I8MJW9 A0A1B0BUZ3 A0A0M5IY18 A0A182GUX7 A0A088ALV6 B4GDM0 A0A1A9WBZ7 A0A1A9Y2J5 A0A0L7QSX8 A0A154PBE0 A0A1A9VJ30 Q16PT8 A0A182M4X4 A0A0J7NWU6 A0A195C6Y5 A0A182R3J2 A0A3B0J4F9 A0A182YML3 E2AVZ5 A0A182WJR8 A0A151X4R7 A0A026X145 E9IGS5 A0A1Q3F680 E0VM14 A0A158NDX0 A0A084WER8 F4W7U5 A0A195BPP4 A0A182JYP7 A0A182N920 W8C3S0 A0A182PP37 A0A182XLJ4 A0A182V082 E2BY63 A0A182KQN5 A0A182JEE3 A0A182UC60 A0A195D7W1 A0A182I2Q9 A0A182QLE3 A0A195ERA4 Q7PT33 W5J8A4 A0A224XI80 A0A023F5M0 B0WVX3 A0A0K8WDI6 A0A034VE32 A0A2M4A9C4 A0A2M4B9Q7 A0A182FGC7 A0A0A9YMY4 A0A336M1Z5 A0A1B0CYN0 A0A0J9RCJ8 A0A1A9Z7A1 K7J1G4 A0A310SB34 A0A2S2R601 J9K579 T1IZH9 A0A232FI38 T1HWH8 A0A131YM49 A0A224YNR5 A0A131Y0N9 A0A2R5LI29 A0A131XCM6 A0A0P5ZPF8 A0A0P5VN56
PDB
6QDV
E-value=0,
Score=3769
Ontologies
GO
GO:0005524
GO:0006996
GO:0004386
GO:0003676
GO:0004842
GO:0016021
GO:0003723
GO:0071013
GO:0000390
GO:0034459
GO:0006325
GO:0016887
GO:0048477
GO:0005681
GO:0005634
GO:0000398
GO:0071011
GO:0000381
GO:0030261
GO:0007419
GO:0004004
GO:0000179
GO:0008026
GO:0008270
GO:0006508
GO:0005622
GO:0007264
GO:0016597
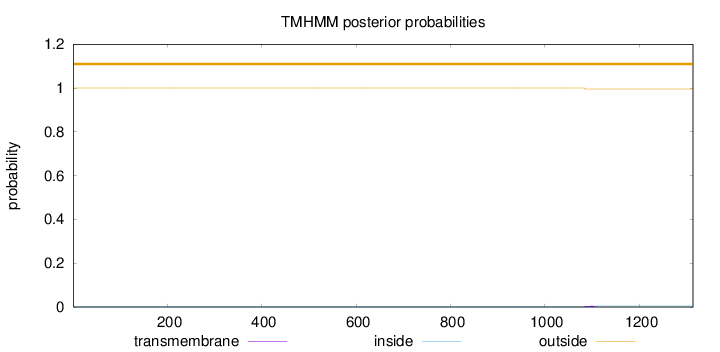
Topology
Subcellular location
Nucleus
Length:
1314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10795
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1314
Population Genetic Test Statistics
Pi
15.898324
Theta
15.58283
Tajima's D
-1.891226
CLR
331.544244
CSRT
0.0196990150492475
Interpretation
Possibly Positive selection
