Gene
KWMTBOMO05968
Pre Gene Modal
BGIBMGA002806
Annotation
PREDICTED:_tRNA_pseudouridine_synthase-like_1_[Bombyx_mori]
Full name
tRNA pseudouridine synthase
Location in the cell
Cytoplasmic Reliability : 2.37
Sequence
CDS
ATGAATAGGTATTTCTTTGTCAACAACATAGGTATAATTGTGAAGCAGTGCCTCAGGGTGTCAGATAATTTTAATGCAAGGTTCAATGCCACTTCCAGGAGATACTTATATAGACTAGCAGTACTAAAAAAAGATGTAGTGATACCAGAGAATGTCTCTATAGCAAATGTCATACCTGTGGAAGAATGGAAACGGTGCCACTTCATCAGATTAGCAGAATTCGACATAGAAAAATTCAAAGAGGGAGCTAAACATTTAGAAGGCTACCACGACTTCTCGACGTTCAAGAAATTCGACAAACTACTCCAACACAAACACAACCGCAGAGAGTTGAAGAGCGTAACAATAAGGGCGGGTAAACCCGTTGTGACTGCGCACACGGAGAGCAGGGGGAGTGCATTCGATTATTGGGATATCGAATTCCACGGACGCGCTTTTGTACATAATCAGATTCGGCGTATGGTAGGCACTCTGATATCAGTAGCCATCGGCAAACTCCAGCCTGACGACGTCAAGGTTATGCTCCAGATACCATCGAAGCACTCGTGGCATTCTTTCATACAGAACTGCCCTTCGGACGGTCTATATCTTTGCGAAGTGGAATACGACCCTAAAGACCTGTTTTACGACCCCGATACTACTGTTGAAAGTGAAACACTCGTTGAATCATAA
Protein
MNRYFFVNNIGIIVKQCLRVSDNFNARFNATSRRYLYRLAVLKKDVVIPENVSIANVIPVEEWKRCHFIRLAEFDIEKFKEGAKHLEGYHDFSTFKKFDKLLQHKHNRRELKSVTIRAGKPVVTAHTESRGSAFDYWDIEFHGRAFVHNQIRRMVGTLISVAIGKLQPDDVKVMLQIPSKHSWHSFIQNCPSDGLYLCEVEYDPKDLFYDPDTTVESETLVES
Summary
Catalytic Activity
tRNA uridine(38-40) = tRNA pseudouridine(38-40)
Similarity
Belongs to the tRNA pseudouridine synthase TruA family.
Uniprot
H9IZX1
A0A194QWL7
A0A194Q466
A0A2H1VKW0
A0A2A4J818
A0A212EIV9
+ More
A0A2M4BU41 A0A2M4BUX4 W5JSH5 A0A034VXN8 A0A1I8PMQ8 A0A182XJK7 A0A2J7PPY8 A0A182HRI5 A0A182KTK0 A0A0L0CQP4 A0A2J7PPY1 A0A0A1WKJ3 A0A0K8TWE1 A0A1A9W4L1 A0A0K8U6C1 A0A0A1XQU0 A0A067RRG6 Q7Q194 B4L909 A0A1I8NK07 A0A1I8M3Y7 A0A0P4VSL0 B4LCD6 A0A0P4W117 A0A1B0A2G3 A0A0M4EJA4 A0A182RR77 A0A1B0FA01 A0A1A9VMW8 A0A182GSG6 Q17EU6 A0A1B0D3R1 B4MXC6 A0A182JTT6 W8BMP8 A0A182NAS1 A0A1A9YN28 B4J2A3 B3M6Q9 A0A0P4VX88 A0A1B0BAB6 Q29EU3 N6TQ04 A0A182W910 A0A0Q5UGL2 B4H5I1 A0A3B0JRM1 A0A182F3V6 A0A182IRN8 A0A0J9RKT0 A0A084W8M8 A0A232FMS0 A0A0R1DU96 Q0E8K6 E2AYB2 A0A182U639 A0A182MJE1 A0A182PTD3 A0A151WME1 K7IPG0 A0A1W4W018 A0A1W4VNI6 A0A1W4W1J5 E9ISQ0 A0A195EVE9 A0A182UUY2 A0A146M7L6 A0A151IAD9 A0A0N5DHI8 B0WVR9 A0A1S4JXH1 A0A088APT9 J9KSS6 B4J2A2 A0A1D1V7L4 A0A158NR53 A0A195B174 A0A2H8TGN9 A0A023F0N7 A0A1D1V9U0 A0A026WIW9 A0A093HXE6 A0A2A3EJX2 M4AWN4 A0A0V0GAZ6 A0A096LSI9 K7F4A5 A0A3B3TRI5 A0A3B3WBX1 A0A336LWW1 A0A0L8FKI5
A0A2M4BU41 A0A2M4BUX4 W5JSH5 A0A034VXN8 A0A1I8PMQ8 A0A182XJK7 A0A2J7PPY8 A0A182HRI5 A0A182KTK0 A0A0L0CQP4 A0A2J7PPY1 A0A0A1WKJ3 A0A0K8TWE1 A0A1A9W4L1 A0A0K8U6C1 A0A0A1XQU0 A0A067RRG6 Q7Q194 B4L909 A0A1I8NK07 A0A1I8M3Y7 A0A0P4VSL0 B4LCD6 A0A0P4W117 A0A1B0A2G3 A0A0M4EJA4 A0A182RR77 A0A1B0FA01 A0A1A9VMW8 A0A182GSG6 Q17EU6 A0A1B0D3R1 B4MXC6 A0A182JTT6 W8BMP8 A0A182NAS1 A0A1A9YN28 B4J2A3 B3M6Q9 A0A0P4VX88 A0A1B0BAB6 Q29EU3 N6TQ04 A0A182W910 A0A0Q5UGL2 B4H5I1 A0A3B0JRM1 A0A182F3V6 A0A182IRN8 A0A0J9RKT0 A0A084W8M8 A0A232FMS0 A0A0R1DU96 Q0E8K6 E2AYB2 A0A182U639 A0A182MJE1 A0A182PTD3 A0A151WME1 K7IPG0 A0A1W4W018 A0A1W4VNI6 A0A1W4W1J5 E9ISQ0 A0A195EVE9 A0A182UUY2 A0A146M7L6 A0A151IAD9 A0A0N5DHI8 B0WVR9 A0A1S4JXH1 A0A088APT9 J9KSS6 B4J2A2 A0A1D1V7L4 A0A158NR53 A0A195B174 A0A2H8TGN9 A0A023F0N7 A0A1D1V9U0 A0A026WIW9 A0A093HXE6 A0A2A3EJX2 M4AWN4 A0A0V0GAZ6 A0A096LSI9 K7F4A5 A0A3B3TRI5 A0A3B3WBX1 A0A336LWW1 A0A0L8FKI5
EC Number
5.4.99.-
Pubmed
19121390
26354079
22118469
20920257
23761445
25348373
+ More
20966253 26108605 25830018 24845553 12364791 17994087 25315136 26483478 17510324 24495485 15632085 23537049 22936249 24438588 28648823 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 20075255 21282665 26823975 27649274 21347285 25474469 24508170 23542700 17381049
20966253 26108605 25830018 24845553 12364791 17994087 25315136 26483478 17510324 24495485 15632085 23537049 22936249 24438588 28648823 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 20075255 21282665 26823975 27649274 21347285 25474469 24508170 23542700 17381049
EMBL
BABH01018982
BABH01018983
KQ461175
KPJ07966.1
KQ459472
KPJ00139.1
+ More
ODYU01003117 SOQ41470.1 NWSH01002508 PCG68111.1 AGBW02014561 OWR41419.1 GGFJ01007428 MBW56569.1 GGFJ01007427 MBW56568.1 ADMH02000223 ETN67327.1 GAKP01012085 JAC46867.1 NEVH01022641 PNF18391.1 APCN01001706 JRES01000036 KNC34695.1 PNF18392.1 GBXI01014950 JAC99341.1 GDHF01033545 GDHF01020423 JAI18769.1 JAI31891.1 GDHF01030208 GDHF01002888 JAI22106.1 JAI49426.1 GBXI01001042 JAD13250.1 KK852510 KDR22354.1 AAAB01008980 EAA14028.4 CH933816 EDW17184.1 GDRN01100818 GDRN01100815 JAI58494.1 CH940647 EDW68781.2 GDRN01100823 GDRN01100819 GDRN01100817 JAI58490.1 CP012525 ALC44076.1 CCAG010020111 JXUM01084971 JXUM01084972 KQ563470 KXJ73784.1 CH477279 EAT45005.1 AJVK01023646 CH963876 EDW76959.1 GAMC01011904 JAB94651.1 CH916366 EDV95962.1 CH902618 EDV38709.1 GDRN01100822 GDRN01100816 JAI58487.1 JXJN01010889 CH379070 EAL29967.3 APGK01056097 KB741274 ENN71335.1 CH954178 KQS43002.1 CH479211 EDW33033.1 OUUW01000009 SPP84784.1 CM002912 KMY96578.1 ATLV01021496 KE525319 KFB46572.1 NNAY01000011 OXU32036.1 CM000159 KRK00645.1 AE014296 BT088434 ABI31225.1 ACR40402.1 GL443864 EFN61571.1 AXCM01008285 KQ982944 KYQ48977.1 GL765434 EFZ16279.1 KQ981965 KYN31877.1 GDHC01003883 JAQ14746.1 KQ978233 KYM96073.1 DS232131 EDS35676.1 ABLF02027850 ABLF02027854 EDV95961.1 BDGG01000004 GAU97689.1 ADTU01023705 KQ976691 KYM77954.1 GFXV01001459 MBW13264.1 GBBI01004161 JAC14551.1 GAU97690.1 KK107182 EZA55987.1 KK399589 KFV59153.1 KZ288232 PBC31486.1 GECL01000881 JAP05243.1 AYCK01016379 AYCK01016380 AGCU01113450 AGCU01113451 AGCU01113452 AGCU01113453 AGCU01113454 AGCU01113455 AGCU01113456 AGCU01113457 UFQT01000257 SSX22454.1 KQ430152 KOF64598.1
ODYU01003117 SOQ41470.1 NWSH01002508 PCG68111.1 AGBW02014561 OWR41419.1 GGFJ01007428 MBW56569.1 GGFJ01007427 MBW56568.1 ADMH02000223 ETN67327.1 GAKP01012085 JAC46867.1 NEVH01022641 PNF18391.1 APCN01001706 JRES01000036 KNC34695.1 PNF18392.1 GBXI01014950 JAC99341.1 GDHF01033545 GDHF01020423 JAI18769.1 JAI31891.1 GDHF01030208 GDHF01002888 JAI22106.1 JAI49426.1 GBXI01001042 JAD13250.1 KK852510 KDR22354.1 AAAB01008980 EAA14028.4 CH933816 EDW17184.1 GDRN01100818 GDRN01100815 JAI58494.1 CH940647 EDW68781.2 GDRN01100823 GDRN01100819 GDRN01100817 JAI58490.1 CP012525 ALC44076.1 CCAG010020111 JXUM01084971 JXUM01084972 KQ563470 KXJ73784.1 CH477279 EAT45005.1 AJVK01023646 CH963876 EDW76959.1 GAMC01011904 JAB94651.1 CH916366 EDV95962.1 CH902618 EDV38709.1 GDRN01100822 GDRN01100816 JAI58487.1 JXJN01010889 CH379070 EAL29967.3 APGK01056097 KB741274 ENN71335.1 CH954178 KQS43002.1 CH479211 EDW33033.1 OUUW01000009 SPP84784.1 CM002912 KMY96578.1 ATLV01021496 KE525319 KFB46572.1 NNAY01000011 OXU32036.1 CM000159 KRK00645.1 AE014296 BT088434 ABI31225.1 ACR40402.1 GL443864 EFN61571.1 AXCM01008285 KQ982944 KYQ48977.1 GL765434 EFZ16279.1 KQ981965 KYN31877.1 GDHC01003883 JAQ14746.1 KQ978233 KYM96073.1 DS232131 EDS35676.1 ABLF02027850 ABLF02027854 EDV95961.1 BDGG01000004 GAU97689.1 ADTU01023705 KQ976691 KYM77954.1 GFXV01001459 MBW13264.1 GBBI01004161 JAC14551.1 GAU97690.1 KK107182 EZA55987.1 KK399589 KFV59153.1 KZ288232 PBC31486.1 GECL01000881 JAP05243.1 AYCK01016379 AYCK01016380 AGCU01113450 AGCU01113451 AGCU01113452 AGCU01113453 AGCU01113454 AGCU01113455 AGCU01113456 AGCU01113457 UFQT01000257 SSX22454.1 KQ430152 KOF64598.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000000673
+ More
UP000095300 UP000076407 UP000235965 UP000075840 UP000075882 UP000037069 UP000091820 UP000027135 UP000007062 UP000009192 UP000095301 UP000008792 UP000092445 UP000092553 UP000075900 UP000092444 UP000078200 UP000069940 UP000249989 UP000008820 UP000092462 UP000007798 UP000075881 UP000075884 UP000092443 UP000001070 UP000007801 UP000092460 UP000001819 UP000019118 UP000075920 UP000008711 UP000008744 UP000268350 UP000069272 UP000075880 UP000030765 UP000215335 UP000002282 UP000000803 UP000000311 UP000075902 UP000075883 UP000075885 UP000075809 UP000002358 UP000192221 UP000078541 UP000075903 UP000078542 UP000046395 UP000002320 UP000005203 UP000007819 UP000186922 UP000005205 UP000078540 UP000053097 UP000242457 UP000002852 UP000028760 UP000007267 UP000261500 UP000261480 UP000053454
UP000095300 UP000076407 UP000235965 UP000075840 UP000075882 UP000037069 UP000091820 UP000027135 UP000007062 UP000009192 UP000095301 UP000008792 UP000092445 UP000092553 UP000075900 UP000092444 UP000078200 UP000069940 UP000249989 UP000008820 UP000092462 UP000007798 UP000075881 UP000075884 UP000092443 UP000001070 UP000007801 UP000092460 UP000001819 UP000019118 UP000075920 UP000008711 UP000008744 UP000268350 UP000069272 UP000075880 UP000030765 UP000215335 UP000002282 UP000000803 UP000000311 UP000075902 UP000075883 UP000075885 UP000075809 UP000002358 UP000192221 UP000078541 UP000075903 UP000078542 UP000046395 UP000002320 UP000005203 UP000007819 UP000186922 UP000005205 UP000078540 UP000053097 UP000242457 UP000002852 UP000028760 UP000007267 UP000261500 UP000261480 UP000053454
Interpro
IPR020095
PsdUridine_synth_TruA_C
+ More
IPR001406 PsdUridine_synth_TruA
IPR020103 PsdUridine_synth_cat_dom_sf
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR003595 Tyr_Pase_cat
IPR017423 TRM6
IPR001406 PsdUridine_synth_TruA
IPR020103 PsdUridine_synth_cat_dom_sf
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR003595 Tyr_Pase_cat
IPR017423 TRM6
Gene 3D
CDD
ProteinModelPortal
H9IZX1
A0A194QWL7
A0A194Q466
A0A2H1VKW0
A0A2A4J818
A0A212EIV9
+ More
A0A2M4BU41 A0A2M4BUX4 W5JSH5 A0A034VXN8 A0A1I8PMQ8 A0A182XJK7 A0A2J7PPY8 A0A182HRI5 A0A182KTK0 A0A0L0CQP4 A0A2J7PPY1 A0A0A1WKJ3 A0A0K8TWE1 A0A1A9W4L1 A0A0K8U6C1 A0A0A1XQU0 A0A067RRG6 Q7Q194 B4L909 A0A1I8NK07 A0A1I8M3Y7 A0A0P4VSL0 B4LCD6 A0A0P4W117 A0A1B0A2G3 A0A0M4EJA4 A0A182RR77 A0A1B0FA01 A0A1A9VMW8 A0A182GSG6 Q17EU6 A0A1B0D3R1 B4MXC6 A0A182JTT6 W8BMP8 A0A182NAS1 A0A1A9YN28 B4J2A3 B3M6Q9 A0A0P4VX88 A0A1B0BAB6 Q29EU3 N6TQ04 A0A182W910 A0A0Q5UGL2 B4H5I1 A0A3B0JRM1 A0A182F3V6 A0A182IRN8 A0A0J9RKT0 A0A084W8M8 A0A232FMS0 A0A0R1DU96 Q0E8K6 E2AYB2 A0A182U639 A0A182MJE1 A0A182PTD3 A0A151WME1 K7IPG0 A0A1W4W018 A0A1W4VNI6 A0A1W4W1J5 E9ISQ0 A0A195EVE9 A0A182UUY2 A0A146M7L6 A0A151IAD9 A0A0N5DHI8 B0WVR9 A0A1S4JXH1 A0A088APT9 J9KSS6 B4J2A2 A0A1D1V7L4 A0A158NR53 A0A195B174 A0A2H8TGN9 A0A023F0N7 A0A1D1V9U0 A0A026WIW9 A0A093HXE6 A0A2A3EJX2 M4AWN4 A0A0V0GAZ6 A0A096LSI9 K7F4A5 A0A3B3TRI5 A0A3B3WBX1 A0A336LWW1 A0A0L8FKI5
A0A2M4BU41 A0A2M4BUX4 W5JSH5 A0A034VXN8 A0A1I8PMQ8 A0A182XJK7 A0A2J7PPY8 A0A182HRI5 A0A182KTK0 A0A0L0CQP4 A0A2J7PPY1 A0A0A1WKJ3 A0A0K8TWE1 A0A1A9W4L1 A0A0K8U6C1 A0A0A1XQU0 A0A067RRG6 Q7Q194 B4L909 A0A1I8NK07 A0A1I8M3Y7 A0A0P4VSL0 B4LCD6 A0A0P4W117 A0A1B0A2G3 A0A0M4EJA4 A0A182RR77 A0A1B0FA01 A0A1A9VMW8 A0A182GSG6 Q17EU6 A0A1B0D3R1 B4MXC6 A0A182JTT6 W8BMP8 A0A182NAS1 A0A1A9YN28 B4J2A3 B3M6Q9 A0A0P4VX88 A0A1B0BAB6 Q29EU3 N6TQ04 A0A182W910 A0A0Q5UGL2 B4H5I1 A0A3B0JRM1 A0A182F3V6 A0A182IRN8 A0A0J9RKT0 A0A084W8M8 A0A232FMS0 A0A0R1DU96 Q0E8K6 E2AYB2 A0A182U639 A0A182MJE1 A0A182PTD3 A0A151WME1 K7IPG0 A0A1W4W018 A0A1W4VNI6 A0A1W4W1J5 E9ISQ0 A0A195EVE9 A0A182UUY2 A0A146M7L6 A0A151IAD9 A0A0N5DHI8 B0WVR9 A0A1S4JXH1 A0A088APT9 J9KSS6 B4J2A2 A0A1D1V7L4 A0A158NR53 A0A195B174 A0A2H8TGN9 A0A023F0N7 A0A1D1V9U0 A0A026WIW9 A0A093HXE6 A0A2A3EJX2 M4AWN4 A0A0V0GAZ6 A0A096LSI9 K7F4A5 A0A3B3TRI5 A0A3B3WBX1 A0A336LWW1 A0A0L8FKI5
PDB
1VS3
E-value=3.35996e-08,
Score=136
Ontologies
GO
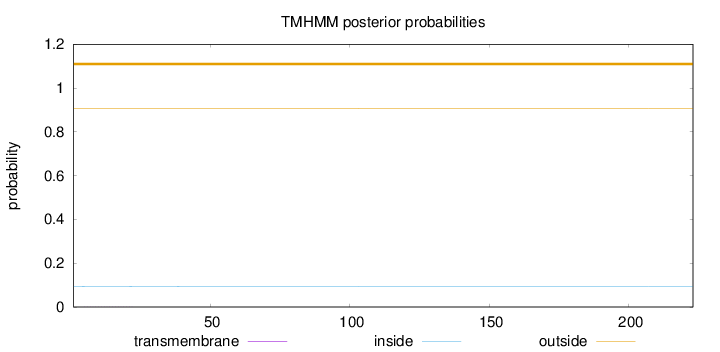
Topology
Length:
223
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00757999999999999
Exp number, first 60 AAs:
0.00613
Total prob of N-in:
0.09340
outside
1 - 223
Population Genetic Test Statistics
Pi
17.725855
Theta
15.337031
Tajima's D
0.557391
CLR
0.333287
CSRT
0.543872806359682
Interpretation
Uncertain
