Pre Gene Modal
BGIBMGA002898
Annotation
PREDICTED:_transmembrane_emp24_domain-containing_protein_5-like_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 2.123
Sequence
CDS
ATGGATTACAAAGTCCACCTCGATGCTGGGAAGGAAGATTGTTACTTTCAATATGTTCAACCCGGTGCCACTTTCTACGCCAGTTATCAGGTGTTGAAAGGAGGTGACGGCATGTGCGGGTTCGCTGTCCGTCATCCAAATGGCAAAATAGTTCATCCATATGAATGGAGACAAAGTGCAGAGTATGCTGATCAATCATCATCAGGCGGCTACTATGCTGTGTGTATTGACAACCAATTCTCAAGGTTTGCTGGAAAATTAGTTAATCTGTACTTGTCGGTGATCAGATACGATATGTGGGAGAAGTATGCGAAAGAGGTTGAGGAACTGGACATGAATATCAAAAATTTTACTTCTTCCATTCAGTTTGTGGAGAGGAACATTAATGACATACTGCAGTACCAGTATCACTCGAGGGCGAGAGAATCTCGTGATTACAACCTGCTGTTAGACAACAATATTTATGTACTTAGTTAA
Protein
MDYKVHLDAGKEDCYFQYVQPGATFYASYQVLKGGDGMCGFAVRHPNGKIVHPYEWRQSAEYADQSSSGGYYAVCIDNQFSRFAGKLVNLYLSVIRYDMWEKYAKEVEELDMNIKNFTSSIQFVERNINDILQYQYHSRARESRDYNLLLDNNIYVLS
Summary
Similarity
Belongs to the EMP24/GP25L family.
Uniprot
H9J063
A0A2H1VKZ6
I4DK31
A0A2A4J9H4
A0A194QSK6
A0A212EIX9
+ More
S4P956 A0A0L7KP74 A0A1L8DVH5 D6WZ79 A0A0T6AWN0 A0A2J7PJI6 A0A1Y1K4L5 A0A067RMQ6 A0A0A9XVR9 A0A0L0BTV8 A0A1W4W4P6 A0A1S3CZ35 A0A1I8P282 A0A1B6MKD3 A0A1L8EAX0 A0A1B0FQU4 A0A1A9VHB6 A0A1A9Z952 A0A1A9XR20 A0A1B0BRQ8 B4LHE2 A0A1B6GFZ1 A0A1B6JVJ4 N6SYY8 B0VZH6 A0A1Q3FBU9 U5ES02 R4G3Q9 A0A0M4EZ45 A0A0M4EJM0 A0A2R7WX93 A0A2S2Q3D4 T1PCF7 A0A151X2R5 B4KWP9 B3MA32 A0A1I8MPH8 E2C030 A0A224XVZ4 A0A3B0KJ45 B4MKZ1 C4WS48 B4HUP2 A0A3B0KAG0 A0A2H8THS3 Q9VRU2 A0A0J9RQ46 W8C511 A0A195E429 A0A1W4VA47 A0A158P2A1 B3NGD2 A0A195FJY4 F4W6J4 A0A0Q5UIE7 B4PJC8 A0A151I074 A0A195D349 A0A1A9W4W0 B4H6G0 Q2LZD7 A0A0A1WYA9 K7J2P2 B4QJ94 B4IZS6 A0A182NKF2 Q175W1 A0A182SUR6 A0A023EL11 A0A1J1HEF9 A0A182GX41 A0A026WE64 A0A0K8UT07 A0A182M7H0 A0A182UWV1 A0A182UH91 A0A182QNS8 A0A0C9QHG9 A0A2M4AH42 A0A2M4BZC0 Q5TR78 A0A182FE27 A0A182PJT7 E9IC16 E0VAV3 E2AUA2 A0A182I1N1 A0A0M9A9K7 A0A2A3EL77 A0A084W433 A0A087ZW69 A0A1Y9IVF1 A0A2M3ZE76 A0A336LF25
S4P956 A0A0L7KP74 A0A1L8DVH5 D6WZ79 A0A0T6AWN0 A0A2J7PJI6 A0A1Y1K4L5 A0A067RMQ6 A0A0A9XVR9 A0A0L0BTV8 A0A1W4W4P6 A0A1S3CZ35 A0A1I8P282 A0A1B6MKD3 A0A1L8EAX0 A0A1B0FQU4 A0A1A9VHB6 A0A1A9Z952 A0A1A9XR20 A0A1B0BRQ8 B4LHE2 A0A1B6GFZ1 A0A1B6JVJ4 N6SYY8 B0VZH6 A0A1Q3FBU9 U5ES02 R4G3Q9 A0A0M4EZ45 A0A0M4EJM0 A0A2R7WX93 A0A2S2Q3D4 T1PCF7 A0A151X2R5 B4KWP9 B3MA32 A0A1I8MPH8 E2C030 A0A224XVZ4 A0A3B0KJ45 B4MKZ1 C4WS48 B4HUP2 A0A3B0KAG0 A0A2H8THS3 Q9VRU2 A0A0J9RQ46 W8C511 A0A195E429 A0A1W4VA47 A0A158P2A1 B3NGD2 A0A195FJY4 F4W6J4 A0A0Q5UIE7 B4PJC8 A0A151I074 A0A195D349 A0A1A9W4W0 B4H6G0 Q2LZD7 A0A0A1WYA9 K7J2P2 B4QJ94 B4IZS6 A0A182NKF2 Q175W1 A0A182SUR6 A0A023EL11 A0A1J1HEF9 A0A182GX41 A0A026WE64 A0A0K8UT07 A0A182M7H0 A0A182UWV1 A0A182UH91 A0A182QNS8 A0A0C9QHG9 A0A2M4AH42 A0A2M4BZC0 Q5TR78 A0A182FE27 A0A182PJT7 E9IC16 E0VAV3 E2AUA2 A0A182I1N1 A0A0M9A9K7 A0A2A3EL77 A0A084W433 A0A087ZW69 A0A1Y9IVF1 A0A2M3ZE76 A0A336LF25
Pubmed
19121390
22651552
26354079
22118469
23622113
26227816
+ More
18362917 19820115 28004739 24845553 25401762 26823975 26108605 17994087 23537049 18057021 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24495485 21347285 21719571 17550304 15632085 25830018 20075255 17510324 24945155 26483478 24508170 30249741 12364791 21282665 20566863 24438588
18362917 19820115 28004739 24845553 25401762 26823975 26108605 17994087 23537049 18057021 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24495485 21347285 21719571 17550304 15632085 25830018 20075255 17510324 24945155 26483478 24508170 30249741 12364791 21282665 20566863 24438588
EMBL
BABH01018984
ODYU01003117
SOQ41471.1
AK401649
KQ459472
BAM18271.1
+ More
KPJ00138.1 NWSH01002508 PCG68110.1 KQ461175 KPJ07965.1 AGBW02014561 OWR41420.1 GAIX01005671 JAA86889.1 JTDY01007905 KOB64866.1 GFDF01003643 JAV10441.1 KQ971372 EFA10393.1 LJIG01022687 KRT79253.1 NEVH01024947 PNF16493.1 GEZM01092974 JAV56402.1 KK852575 KDR21000.1 GBHO01031597 GBHO01031509 GBHO01020128 GBHO01011041 GBRD01015487 GBRD01015486 GBRD01015484 GBRD01015483 GBRD01015482 GBRD01015481 GBRD01015480 GBRD01001913 GDHC01020635 GDHC01005514 JAG12007.1 JAG12095.1 JAG23476.1 JAG32563.1 JAG50339.1 JAP97993.1 JAQ13115.1 JRES01001347 KNC23515.1 GEBQ01003603 JAT36374.1 GFDG01002902 JAV15897.1 CCAG010003582 JXJN01019236 CH940647 EDW68472.1 GECZ01008439 JAS61330.1 GECU01004507 JAT03200.1 APGK01059180 APGK01059181 KB741292 KB631594 ENN70453.1 ERL84521.1 DS231814 EDS31713.1 GFDL01010056 JAV24989.1 GANO01003471 JAB56400.1 ACPB03020838 GAHY01002005 JAA75505.1 CP012525 ALC43595.1 ALC43210.1 KK855844 PTY24081.1 GGMS01003013 MBY72216.1 KA645815 AFP60444.1 KQ982580 KYQ54494.1 CH933809 EDW17496.1 CH902618 EDV41252.1 KPU79010.1 GL451712 EFN78726.1 GFTR01003786 JAW12640.1 OUUW01000009 SPP85111.1 CH963847 EDW72916.1 ABLF02014852 AK340045 BAH70718.1 CH480817 EDW50663.1 SPP85110.1 GFXV01001878 MBW13683.1 AE014296 AY061384 AAF50698.2 AAL28932.1 AAN12236.2 AAN12237.2 CM002912 KMY97862.1 GAMC01002047 JAC04509.1 KQ979657 KYN19940.1 ADTU01007043 CH954178 EDV51098.2 KQS43671.1 KQ981523 KYN40314.1 GL887707 EGI70329.1 KQS43670.1 CM000159 EDW93596.2 KRK01264.1 KQ976632 KYM78888.1 KQ976885 KYN07291.1 CH479214 EDW33388.1 CH379069 EAL29571.3 GBXI01010909 JAD03383.1 CM000363 EDX09403.1 CH916366 EDV95661.1 CH477395 EAT41842.1 GAPW01003705 JAC09893.1 CVRI01000001 CRK86219.1 JXUM01094624 JXUM01094625 KQ564141 KXJ72691.1 KK107250 QOIP01000002 EZA54357.1 RLU25485.1 GDHF01022603 JAI29711.1 AXCM01004994 AXCN02000400 GBYB01014083 JAG83850.1 GGFK01006788 MBW40109.1 GGFJ01009221 MBW58362.1 AAAB01008960 EAL39986.3 GL762150 EFZ21886.1 DS235016 EEB10509.1 GL442815 EFN62959.1 APCN01000070 KQ435713 KOX79286.1 KZ288215 PBC32467.1 ATLV01020271 KE525297 KFB44977.1 GGFM01006030 MBW26781.1 UFQS01000200 UFQS01003214 UFQT01000200 UFQT01003214 SSX01237.1 SSX15367.1 SSX21617.1
KPJ00138.1 NWSH01002508 PCG68110.1 KQ461175 KPJ07965.1 AGBW02014561 OWR41420.1 GAIX01005671 JAA86889.1 JTDY01007905 KOB64866.1 GFDF01003643 JAV10441.1 KQ971372 EFA10393.1 LJIG01022687 KRT79253.1 NEVH01024947 PNF16493.1 GEZM01092974 JAV56402.1 KK852575 KDR21000.1 GBHO01031597 GBHO01031509 GBHO01020128 GBHO01011041 GBRD01015487 GBRD01015486 GBRD01015484 GBRD01015483 GBRD01015482 GBRD01015481 GBRD01015480 GBRD01001913 GDHC01020635 GDHC01005514 JAG12007.1 JAG12095.1 JAG23476.1 JAG32563.1 JAG50339.1 JAP97993.1 JAQ13115.1 JRES01001347 KNC23515.1 GEBQ01003603 JAT36374.1 GFDG01002902 JAV15897.1 CCAG010003582 JXJN01019236 CH940647 EDW68472.1 GECZ01008439 JAS61330.1 GECU01004507 JAT03200.1 APGK01059180 APGK01059181 KB741292 KB631594 ENN70453.1 ERL84521.1 DS231814 EDS31713.1 GFDL01010056 JAV24989.1 GANO01003471 JAB56400.1 ACPB03020838 GAHY01002005 JAA75505.1 CP012525 ALC43595.1 ALC43210.1 KK855844 PTY24081.1 GGMS01003013 MBY72216.1 KA645815 AFP60444.1 KQ982580 KYQ54494.1 CH933809 EDW17496.1 CH902618 EDV41252.1 KPU79010.1 GL451712 EFN78726.1 GFTR01003786 JAW12640.1 OUUW01000009 SPP85111.1 CH963847 EDW72916.1 ABLF02014852 AK340045 BAH70718.1 CH480817 EDW50663.1 SPP85110.1 GFXV01001878 MBW13683.1 AE014296 AY061384 AAF50698.2 AAL28932.1 AAN12236.2 AAN12237.2 CM002912 KMY97862.1 GAMC01002047 JAC04509.1 KQ979657 KYN19940.1 ADTU01007043 CH954178 EDV51098.2 KQS43671.1 KQ981523 KYN40314.1 GL887707 EGI70329.1 KQS43670.1 CM000159 EDW93596.2 KRK01264.1 KQ976632 KYM78888.1 KQ976885 KYN07291.1 CH479214 EDW33388.1 CH379069 EAL29571.3 GBXI01010909 JAD03383.1 CM000363 EDX09403.1 CH916366 EDV95661.1 CH477395 EAT41842.1 GAPW01003705 JAC09893.1 CVRI01000001 CRK86219.1 JXUM01094624 JXUM01094625 KQ564141 KXJ72691.1 KK107250 QOIP01000002 EZA54357.1 RLU25485.1 GDHF01022603 JAI29711.1 AXCM01004994 AXCN02000400 GBYB01014083 JAG83850.1 GGFK01006788 MBW40109.1 GGFJ01009221 MBW58362.1 AAAB01008960 EAL39986.3 GL762150 EFZ21886.1 DS235016 EEB10509.1 GL442815 EFN62959.1 APCN01000070 KQ435713 KOX79286.1 KZ288215 PBC32467.1 ATLV01020271 KE525297 KFB44977.1 GGFM01006030 MBW26781.1 UFQS01000200 UFQS01003214 UFQT01000200 UFQT01003214 SSX01237.1 SSX15367.1 SSX21617.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000235965 UP000027135 UP000037069 UP000192223 UP000079169 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000008792 UP000019118 UP000030742 UP000002320 UP000015103 UP000092553 UP000075809 UP000009192 UP000007801 UP000095301 UP000008237 UP000268350 UP000007798 UP000007819 UP000001292 UP000000803 UP000078492 UP000192221 UP000005205 UP000008711 UP000078541 UP000007755 UP000002282 UP000078540 UP000078542 UP000091820 UP000008744 UP000001819 UP000002358 UP000000304 UP000001070 UP000075884 UP000008820 UP000075901 UP000183832 UP000069940 UP000249989 UP000053097 UP000279307 UP000075883 UP000075903 UP000075902 UP000075886 UP000007062 UP000069272 UP000075885 UP000009046 UP000000311 UP000075840 UP000053105 UP000242457 UP000030765 UP000005203 UP000075920
UP000007266 UP000235965 UP000027135 UP000037069 UP000192223 UP000079169 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000008792 UP000019118 UP000030742 UP000002320 UP000015103 UP000092553 UP000075809 UP000009192 UP000007801 UP000095301 UP000008237 UP000268350 UP000007798 UP000007819 UP000001292 UP000000803 UP000078492 UP000192221 UP000005205 UP000008711 UP000078541 UP000007755 UP000002282 UP000078540 UP000078542 UP000091820 UP000008744 UP000001819 UP000002358 UP000000304 UP000001070 UP000075884 UP000008820 UP000075901 UP000183832 UP000069940 UP000249989 UP000053097 UP000279307 UP000075883 UP000075903 UP000075902 UP000075886 UP000007062 UP000069272 UP000075885 UP000009046 UP000000311 UP000075840 UP000053105 UP000242457 UP000030765 UP000005203 UP000075920
Pfam
PF01105 EMP24_GP25L
SUPFAM
SSF101576
SSF101576
ProteinModelPortal
H9J063
A0A2H1VKZ6
I4DK31
A0A2A4J9H4
A0A194QSK6
A0A212EIX9
+ More
S4P956 A0A0L7KP74 A0A1L8DVH5 D6WZ79 A0A0T6AWN0 A0A2J7PJI6 A0A1Y1K4L5 A0A067RMQ6 A0A0A9XVR9 A0A0L0BTV8 A0A1W4W4P6 A0A1S3CZ35 A0A1I8P282 A0A1B6MKD3 A0A1L8EAX0 A0A1B0FQU4 A0A1A9VHB6 A0A1A9Z952 A0A1A9XR20 A0A1B0BRQ8 B4LHE2 A0A1B6GFZ1 A0A1B6JVJ4 N6SYY8 B0VZH6 A0A1Q3FBU9 U5ES02 R4G3Q9 A0A0M4EZ45 A0A0M4EJM0 A0A2R7WX93 A0A2S2Q3D4 T1PCF7 A0A151X2R5 B4KWP9 B3MA32 A0A1I8MPH8 E2C030 A0A224XVZ4 A0A3B0KJ45 B4MKZ1 C4WS48 B4HUP2 A0A3B0KAG0 A0A2H8THS3 Q9VRU2 A0A0J9RQ46 W8C511 A0A195E429 A0A1W4VA47 A0A158P2A1 B3NGD2 A0A195FJY4 F4W6J4 A0A0Q5UIE7 B4PJC8 A0A151I074 A0A195D349 A0A1A9W4W0 B4H6G0 Q2LZD7 A0A0A1WYA9 K7J2P2 B4QJ94 B4IZS6 A0A182NKF2 Q175W1 A0A182SUR6 A0A023EL11 A0A1J1HEF9 A0A182GX41 A0A026WE64 A0A0K8UT07 A0A182M7H0 A0A182UWV1 A0A182UH91 A0A182QNS8 A0A0C9QHG9 A0A2M4AH42 A0A2M4BZC0 Q5TR78 A0A182FE27 A0A182PJT7 E9IC16 E0VAV3 E2AUA2 A0A182I1N1 A0A0M9A9K7 A0A2A3EL77 A0A084W433 A0A087ZW69 A0A1Y9IVF1 A0A2M3ZE76 A0A336LF25
S4P956 A0A0L7KP74 A0A1L8DVH5 D6WZ79 A0A0T6AWN0 A0A2J7PJI6 A0A1Y1K4L5 A0A067RMQ6 A0A0A9XVR9 A0A0L0BTV8 A0A1W4W4P6 A0A1S3CZ35 A0A1I8P282 A0A1B6MKD3 A0A1L8EAX0 A0A1B0FQU4 A0A1A9VHB6 A0A1A9Z952 A0A1A9XR20 A0A1B0BRQ8 B4LHE2 A0A1B6GFZ1 A0A1B6JVJ4 N6SYY8 B0VZH6 A0A1Q3FBU9 U5ES02 R4G3Q9 A0A0M4EZ45 A0A0M4EJM0 A0A2R7WX93 A0A2S2Q3D4 T1PCF7 A0A151X2R5 B4KWP9 B3MA32 A0A1I8MPH8 E2C030 A0A224XVZ4 A0A3B0KJ45 B4MKZ1 C4WS48 B4HUP2 A0A3B0KAG0 A0A2H8THS3 Q9VRU2 A0A0J9RQ46 W8C511 A0A195E429 A0A1W4VA47 A0A158P2A1 B3NGD2 A0A195FJY4 F4W6J4 A0A0Q5UIE7 B4PJC8 A0A151I074 A0A195D349 A0A1A9W4W0 B4H6G0 Q2LZD7 A0A0A1WYA9 K7J2P2 B4QJ94 B4IZS6 A0A182NKF2 Q175W1 A0A182SUR6 A0A023EL11 A0A1J1HEF9 A0A182GX41 A0A026WE64 A0A0K8UT07 A0A182M7H0 A0A182UWV1 A0A182UH91 A0A182QNS8 A0A0C9QHG9 A0A2M4AH42 A0A2M4BZC0 Q5TR78 A0A182FE27 A0A182PJT7 E9IC16 E0VAV3 E2AUA2 A0A182I1N1 A0A0M9A9K7 A0A2A3EL77 A0A084W433 A0A087ZW69 A0A1Y9IVF1 A0A2M3ZE76 A0A336LF25
PDB
5GU5
E-value=1.37766e-06,
Score=119
Ontologies
GO
PANTHER
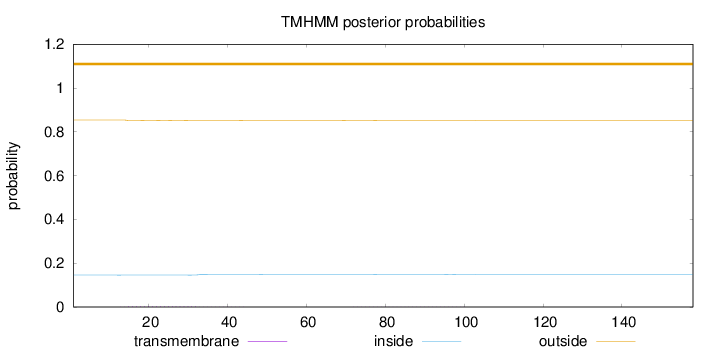
Topology
Subcellular location
Membrane
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0812
Exp number, first 60 AAs:
0.06282
Total prob of N-in:
0.14597
outside
1 - 158
Population Genetic Test Statistics
Pi
17.726814
Theta
18.875389
Tajima's D
-0.444277
CLR
0.875801
CSRT
0.255987200639968
Interpretation
Uncertain
