Gene
KWMTBOMO05954
Pre Gene Modal
BGIBMGA002903
Annotation
PREDICTED:_Bardet-Biedl_syndrome_1_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.499 PlasmaMembrane Reliability : 1.493
Sequence
CDS
ATGACCAGTATGTCCCGCTGGTTGGACGTGGAAGTAGGTACAGATGATATAGATCTAAATACTTTGCCGTCTAATTTAGCTCTTGTCGATCTGCAGAATGATAAAGATTATAAACTACTGGTCGCTGATTTTGGAAAAGGCGAAGAGGGCCCTAAACTTAAGGTATTCAAAGGAACAATGCAAACATCAGACATCCTGTTACCAGAACTACCCCTTGGCATAGTTGGTTTCTATACGTCAGAGGCAATTCCAAGGTCCATGCCAATCATAGCTGTGGCATTCAGCTCTGCAGTATACTTATACAGAAATATGAAGTTTTTTTACAAATATCTATTGCCATCGGTTGATTTGAGCGTCCACGAGATGGATGTTTGGAAGCAGCTAATAAACTCAGAAAATCAGACACCAGAAATGATAACAATACTCCAAGAGAATTTAAAAGCTATTCCTCCGAAGATATTAAGTCTGCAATCTCAGAATTTCCTGGCTCTTACCTTAGACCAGCAGTTGGAATACCTAGAACAAGCTGCCGAACTACCTATGTGGAAACCTCCTGAGATCACATGCATCAGTACATTGAAGATGAACTCAATAGACCAGTATTCTGTGAGTTGTTTGGTAGTCGGTACAGAAGATGGTTACATTATTATTTTGGATCCCCAAACATTTACAACTCTGTCGCAGGGAGAAATCAAGGTCATCACCGCCGATGAAAGCATCATAGTTATATGTACAGACAACACGATGTCATGTTACACCAAAAAGGGTAAGAAGCAGTGGTGCGTCAACTTGGAGCACAGGCCGATATCGATGACTCTGGTGCCGGTACTGCATTTGGGAGTCACCTTAGTGGGGGTGGCGCTGACTTCAGGGCATGTCCACTTCTATGAAGGGAAAGCGCGTTTGGACACATTGTTTGTTAGAGATGTCGCTTCGGTGATGAGGTTCGGTCGGTTGGGTCAAGAGGAGCACGTGCTCATCGTTGTGACCGGCAGTGAGTACATTTTGTAA
Protein
MTSMSRWLDVEVGTDDIDLNTLPSNLALVDLQNDKDYKLLVADFGKGEEGPKLKVFKGTMQTSDILLPELPLGIVGFYTSEAIPRSMPIIAVAFSSAVYLYRNMKFFYKYLLPSVDLSVHEMDVWKQLINSENQTPEMITILQENLKAIPPKILSLQSQNFLALTLDQQLEYLEQAAELPMWKPPEITCISTLKMNSIDQYSVSCLVVGTEDGYIIILDPQTFTTLSQGEIKVITADESIIVICTDNTMSCYTKKGKKQWCVNLEHRPISMTLVPVLHLGVTLVGVALTSGHVHFYEGKARLDTLFVRDVASVMRFGRLGQEEHVLIVVTGSEYIL
Summary
Uniprot
H9J068
A0A2A4JB20
A0A2H1W376
A0A2A4JCJ0
A0A3S2TTQ4
A0A194Q555
+ More
A0A194QWL4 A0A0L7LQI9 B0WXE2 A0A1S4FLT6 Q16WI6 A0A182RSM4 W5JPA7 Q7QJD7 A0A182KRA5 A0A182GWW1 A0A182PQG2 A0A182NHW6 A0A182FF33 A0A336KP14 A0A084WN87 A0A182U0L2 A0A2C9GRU4 A0A182XYV1 A0A182X3S4 A0A182M5X6 A0A182WJ93 A0A182JY52 A0A067RAV1 T1J8I6 A0A1W4X9F6 E0VBP2 A0A182QKI4 A7S5W5 A0A2I4CFM7 T1JYJ2 A0A1B6LXZ3 D6WZ82 A0A1A8IQ14 A0A1A8H3R4 A0A1A8CL89 A0A1A8U323 N6TQ64 A0A3B5LHV2 A0A1A7XV17 A0A1A8Q282 F7DYZ6 A0A1A8MD68 A0A315VHN2 M3ZLB9 B1H2V5 A0A2B4SD42 Q4SE99 U4UBI2 H3CYE9 A0A3Q3AUL7 A0A087YHA2 A0A0S7GH77 A4QP86 K7F525 A0A3Q2XF18 A0A0S7GM05 A0A3M6TAC9 A0A3B3YNA7 A0A0P4VPS7 A0A1B6E6Y3 B3DFT2 A0A3B4GJB6 A0A3B1ITQ8 A0A3Q2WC54 A0A1L8GK24 G3P7I9 A0A3P8Q4B4 A0A3P9CLH6 A0A3P9MWJ2 A0A3B3RF69 W5ULG6 A0A3S2NXZ5 A0A147AID1 A0A3B3BFI8 A0A3Q2CAW1 A0A1S3FK75 A0A3Q3WIM6 A0A3Q3GPE1 S4RMU3 A0A3Q0SEI3 A0A3Q3LZA8 E2BCF4 H2N1F5 A0A3Q1BHR1 A0A3P8SPG8 A0A0K2T5I1 A0A3P9HFF4 A0A1S3I1L6 H2SDM2 A0A3Q1IP34 A0A0M8ZZ34 A0A3P8Z8W1 A0A2U9CQV5 V9KUN1
A0A194QWL4 A0A0L7LQI9 B0WXE2 A0A1S4FLT6 Q16WI6 A0A182RSM4 W5JPA7 Q7QJD7 A0A182KRA5 A0A182GWW1 A0A182PQG2 A0A182NHW6 A0A182FF33 A0A336KP14 A0A084WN87 A0A182U0L2 A0A2C9GRU4 A0A182XYV1 A0A182X3S4 A0A182M5X6 A0A182WJ93 A0A182JY52 A0A067RAV1 T1J8I6 A0A1W4X9F6 E0VBP2 A0A182QKI4 A7S5W5 A0A2I4CFM7 T1JYJ2 A0A1B6LXZ3 D6WZ82 A0A1A8IQ14 A0A1A8H3R4 A0A1A8CL89 A0A1A8U323 N6TQ64 A0A3B5LHV2 A0A1A7XV17 A0A1A8Q282 F7DYZ6 A0A1A8MD68 A0A315VHN2 M3ZLB9 B1H2V5 A0A2B4SD42 Q4SE99 U4UBI2 H3CYE9 A0A3Q3AUL7 A0A087YHA2 A0A0S7GH77 A4QP86 K7F525 A0A3Q2XF18 A0A0S7GM05 A0A3M6TAC9 A0A3B3YNA7 A0A0P4VPS7 A0A1B6E6Y3 B3DFT2 A0A3B4GJB6 A0A3B1ITQ8 A0A3Q2WC54 A0A1L8GK24 G3P7I9 A0A3P8Q4B4 A0A3P9CLH6 A0A3P9MWJ2 A0A3B3RF69 W5ULG6 A0A3S2NXZ5 A0A147AID1 A0A3B3BFI8 A0A3Q2CAW1 A0A1S3FK75 A0A3Q3WIM6 A0A3Q3GPE1 S4RMU3 A0A3Q0SEI3 A0A3Q3LZA8 E2BCF4 H2N1F5 A0A3Q1BHR1 A0A3P8SPG8 A0A0K2T5I1 A0A3P9HFF4 A0A1S3I1L6 H2SDM2 A0A3Q1IP34 A0A0M8ZZ34 A0A3P8Z8W1 A0A2U9CQV5 V9KUN1
Pubmed
19121390
26354079
26227816
17510324
20920257
23761445
+ More
12364791 14747013 17210077 20966253 26483478 24438588 25244985 24845553 20566863 17615350 18362917 19820115 23537049 20431018 29703783 23542700 15496914 17381049 30382153 23594743 25329095 27762356 25186727 29240929 23127152 29451363 20798317 17554307 21551351 25069045 24402279
12364791 14747013 17210077 20966253 26483478 24438588 25244985 24845553 20566863 17615350 18362917 19820115 23537049 20431018 29703783 23542700 15496914 17381049 30382153 23594743 25329095 27762356 25186727 29240929 23127152 29451363 20798317 17554307 21551351 25069045 24402279
EMBL
BABH01019007
BABH01019008
NWSH01002062
PCG69275.1
ODYU01006043
SOQ47539.1
+ More
PCG69274.1 RSAL01000002 RVE54987.1 KQ459472 KPJ00135.1 KQ461175 KPJ07961.1 JTDY01000324 KOB77695.1 DS232163 EDS36472.1 CH477565 EAT38940.1 ADMH02000549 ETN65941.1 AAAB01008807 EAA04615.4 JXUM01094165 KQ564108 KXJ72777.1 UFQS01000582 UFQT01000582 SSX05125.1 SSX25486.1 ATLV01024577 KE525352 KFB51681.1 APCN01000203 AXCM01000778 KK852575 KDR20997.1 JH431954 DS235033 EEB10798.1 AXCN02000381 DS469585 EDO40876.1 CAEY01001108 GEBQ01011420 JAT28557.1 KQ971372 EFA10394.1 HAED01012970 HAEE01008185 SBQ99392.1 HAEB01008917 HAEC01009678 SBQ77894.1 HADZ01016563 HAEA01007120 SBP80504.1 HADY01010838 HAEJ01001051 SBS41508.1 APGK01059190 KB741292 ENN70456.1 HADW01020567 HADX01006423 SBP21967.1 HAEH01009611 HAEI01012939 SBR87601.1 AAMC01091536 HAEF01013327 HAEG01000269 SBR54486.1 NHOQ01001897 PWA21793.1 BC161145 AAI61145.1 LSMT01000095 PFX27801.1 CAAE01014623 CAG01033.1 KB632186 ERL89688.1 AYCK01026358 AYCK01026359 AYCK01026360 GBYX01453111 GBYX01453109 JAO28406.1 BC139697 BC162522 AAI39698.1 AAI62522.1 AGCU01130984 GBYX01453113 GBYX01453108 JAO28402.1 RCHS01004038 RMX38224.1 GDRN01108113 JAI57309.1 GEDC01003605 JAS33693.1 BX511175 BX530023 CR381637 BC162155 AAI62155.1 CM004472 OCT84146.1 JT418748 AHH42831.1 CM012455 RVE59285.1 GCES01008122 JAR78201.1 GL447257 EFN86690.1 HACA01003350 CDW20711.1 KQ435794 KOX73847.1 CP026260 AWP18219.1 JW870158 AFP02676.1
PCG69274.1 RSAL01000002 RVE54987.1 KQ459472 KPJ00135.1 KQ461175 KPJ07961.1 JTDY01000324 KOB77695.1 DS232163 EDS36472.1 CH477565 EAT38940.1 ADMH02000549 ETN65941.1 AAAB01008807 EAA04615.4 JXUM01094165 KQ564108 KXJ72777.1 UFQS01000582 UFQT01000582 SSX05125.1 SSX25486.1 ATLV01024577 KE525352 KFB51681.1 APCN01000203 AXCM01000778 KK852575 KDR20997.1 JH431954 DS235033 EEB10798.1 AXCN02000381 DS469585 EDO40876.1 CAEY01001108 GEBQ01011420 JAT28557.1 KQ971372 EFA10394.1 HAED01012970 HAEE01008185 SBQ99392.1 HAEB01008917 HAEC01009678 SBQ77894.1 HADZ01016563 HAEA01007120 SBP80504.1 HADY01010838 HAEJ01001051 SBS41508.1 APGK01059190 KB741292 ENN70456.1 HADW01020567 HADX01006423 SBP21967.1 HAEH01009611 HAEI01012939 SBR87601.1 AAMC01091536 HAEF01013327 HAEG01000269 SBR54486.1 NHOQ01001897 PWA21793.1 BC161145 AAI61145.1 LSMT01000095 PFX27801.1 CAAE01014623 CAG01033.1 KB632186 ERL89688.1 AYCK01026358 AYCK01026359 AYCK01026360 GBYX01453111 GBYX01453109 JAO28406.1 BC139697 BC162522 AAI39698.1 AAI62522.1 AGCU01130984 GBYX01453113 GBYX01453108 JAO28402.1 RCHS01004038 RMX38224.1 GDRN01108113 JAI57309.1 GEDC01003605 JAS33693.1 BX511175 BX530023 CR381637 BC162155 AAI62155.1 CM004472 OCT84146.1 JT418748 AHH42831.1 CM012455 RVE59285.1 GCES01008122 JAR78201.1 GL447257 EFN86690.1 HACA01003350 CDW20711.1 KQ435794 KOX73847.1 CP026260 AWP18219.1 JW870158 AFP02676.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
+ More
UP000002320 UP000008820 UP000075900 UP000000673 UP000007062 UP000075882 UP000069940 UP000249989 UP000075885 UP000075884 UP000069272 UP000030765 UP000075902 UP000075840 UP000076408 UP000075883 UP000075920 UP000075881 UP000027135 UP000192223 UP000009046 UP000075886 UP000001593 UP000192220 UP000015104 UP000007266 UP000019118 UP000261380 UP000008143 UP000002852 UP000225706 UP000030742 UP000007303 UP000264800 UP000028760 UP000007267 UP000264840 UP000275408 UP000261480 UP000000437 UP000261460 UP000018467 UP000186698 UP000007635 UP000265100 UP000265160 UP000242638 UP000261540 UP000221080 UP000261560 UP000265020 UP000081671 UP000261620 UP000261660 UP000245300 UP000261340 UP000261640 UP000008237 UP000001038 UP000257160 UP000265080 UP000265200 UP000085678 UP000005226 UP000265040 UP000053105 UP000265140 UP000246464
UP000002320 UP000008820 UP000075900 UP000000673 UP000007062 UP000075882 UP000069940 UP000249989 UP000075885 UP000075884 UP000069272 UP000030765 UP000075902 UP000075840 UP000076408 UP000075883 UP000075920 UP000075881 UP000027135 UP000192223 UP000009046 UP000075886 UP000001593 UP000192220 UP000015104 UP000007266 UP000019118 UP000261380 UP000008143 UP000002852 UP000225706 UP000030742 UP000007303 UP000264800 UP000028760 UP000007267 UP000264840 UP000275408 UP000261480 UP000000437 UP000261460 UP000018467 UP000186698 UP000007635 UP000265100 UP000265160 UP000242638 UP000261540 UP000221080 UP000261560 UP000265020 UP000081671 UP000261620 UP000261660 UP000245300 UP000261340 UP000261640 UP000008237 UP000001038 UP000257160 UP000265080 UP000265200 UP000085678 UP000005226 UP000265040 UP000053105 UP000265140 UP000246464
PRIDE
Interpro
IPR028784
BBS1
+ More
IPR032728 BBS1_N
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR011044 Quino_amine_DH_bsu
IPR035984 Acyl-CoA-binding_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR000582 Acyl-CoA-binding_protein
IPR022408 Acyl-CoA-binding_prot_CS
IPR032728 BBS1_N
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR011044 Quino_amine_DH_bsu
IPR035984 Acyl-CoA-binding_sf
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR000582 Acyl-CoA-binding_protein
IPR022408 Acyl-CoA-binding_prot_CS
Gene 3D
CDD
ProteinModelPortal
H9J068
A0A2A4JB20
A0A2H1W376
A0A2A4JCJ0
A0A3S2TTQ4
A0A194Q555
+ More
A0A194QWL4 A0A0L7LQI9 B0WXE2 A0A1S4FLT6 Q16WI6 A0A182RSM4 W5JPA7 Q7QJD7 A0A182KRA5 A0A182GWW1 A0A182PQG2 A0A182NHW6 A0A182FF33 A0A336KP14 A0A084WN87 A0A182U0L2 A0A2C9GRU4 A0A182XYV1 A0A182X3S4 A0A182M5X6 A0A182WJ93 A0A182JY52 A0A067RAV1 T1J8I6 A0A1W4X9F6 E0VBP2 A0A182QKI4 A7S5W5 A0A2I4CFM7 T1JYJ2 A0A1B6LXZ3 D6WZ82 A0A1A8IQ14 A0A1A8H3R4 A0A1A8CL89 A0A1A8U323 N6TQ64 A0A3B5LHV2 A0A1A7XV17 A0A1A8Q282 F7DYZ6 A0A1A8MD68 A0A315VHN2 M3ZLB9 B1H2V5 A0A2B4SD42 Q4SE99 U4UBI2 H3CYE9 A0A3Q3AUL7 A0A087YHA2 A0A0S7GH77 A4QP86 K7F525 A0A3Q2XF18 A0A0S7GM05 A0A3M6TAC9 A0A3B3YNA7 A0A0P4VPS7 A0A1B6E6Y3 B3DFT2 A0A3B4GJB6 A0A3B1ITQ8 A0A3Q2WC54 A0A1L8GK24 G3P7I9 A0A3P8Q4B4 A0A3P9CLH6 A0A3P9MWJ2 A0A3B3RF69 W5ULG6 A0A3S2NXZ5 A0A147AID1 A0A3B3BFI8 A0A3Q2CAW1 A0A1S3FK75 A0A3Q3WIM6 A0A3Q3GPE1 S4RMU3 A0A3Q0SEI3 A0A3Q3LZA8 E2BCF4 H2N1F5 A0A3Q1BHR1 A0A3P8SPG8 A0A0K2T5I1 A0A3P9HFF4 A0A1S3I1L6 H2SDM2 A0A3Q1IP34 A0A0M8ZZ34 A0A3P8Z8W1 A0A2U9CQV5 V9KUN1
A0A194QWL4 A0A0L7LQI9 B0WXE2 A0A1S4FLT6 Q16WI6 A0A182RSM4 W5JPA7 Q7QJD7 A0A182KRA5 A0A182GWW1 A0A182PQG2 A0A182NHW6 A0A182FF33 A0A336KP14 A0A084WN87 A0A182U0L2 A0A2C9GRU4 A0A182XYV1 A0A182X3S4 A0A182M5X6 A0A182WJ93 A0A182JY52 A0A067RAV1 T1J8I6 A0A1W4X9F6 E0VBP2 A0A182QKI4 A7S5W5 A0A2I4CFM7 T1JYJ2 A0A1B6LXZ3 D6WZ82 A0A1A8IQ14 A0A1A8H3R4 A0A1A8CL89 A0A1A8U323 N6TQ64 A0A3B5LHV2 A0A1A7XV17 A0A1A8Q282 F7DYZ6 A0A1A8MD68 A0A315VHN2 M3ZLB9 B1H2V5 A0A2B4SD42 Q4SE99 U4UBI2 H3CYE9 A0A3Q3AUL7 A0A087YHA2 A0A0S7GH77 A4QP86 K7F525 A0A3Q2XF18 A0A0S7GM05 A0A3M6TAC9 A0A3B3YNA7 A0A0P4VPS7 A0A1B6E6Y3 B3DFT2 A0A3B4GJB6 A0A3B1ITQ8 A0A3Q2WC54 A0A1L8GK24 G3P7I9 A0A3P8Q4B4 A0A3P9CLH6 A0A3P9MWJ2 A0A3B3RF69 W5ULG6 A0A3S2NXZ5 A0A147AID1 A0A3B3BFI8 A0A3Q2CAW1 A0A1S3FK75 A0A3Q3WIM6 A0A3Q3GPE1 S4RMU3 A0A3Q0SEI3 A0A3Q3LZA8 E2BCF4 H2N1F5 A0A3Q1BHR1 A0A3P8SPG8 A0A0K2T5I1 A0A3P9HFF4 A0A1S3I1L6 H2SDM2 A0A3Q1IP34 A0A0M8ZZ34 A0A3P8Z8W1 A0A2U9CQV5 V9KUN1
PDB
4V0O
E-value=4.48451e-27,
Score=301
Ontologies
GO
GO:1905515
GO:0034464
GO:0016021
GO:0000062
GO:0061512
GO:0036064
GO:0005930
GO:0005113
GO:0005119
GO:0005813
GO:0061469
GO:0033339
GO:0008593
GO:0070121
GO:0060028
GO:0001947
GO:0060972
GO:0003341
GO:0032402
GO:0060027
GO:0007369
GO:0060026
GO:0060271
GO:0007368
GO:0042445
GO:0051219
GO:0060296
GO:0001764
GO:0008104
GO:0045444
GO:0030534
GO:0021756
GO:0021987
GO:0042048
GO:0001895
GO:0016358
GO:0021766
GO:0061351
GO:0048854
GO:0021591
GO:0007608
GO:0005102
GO:0031514
GO:0044255
GO:0034451
GO:0009566
GO:0051216
GO:0000226
GO:0008594
GO:0005515
GO:0046872
GO:0004784
GO:0005525
GO:0030131
GO:0005488
GO:0005622
GO:0007264
GO:0008026
GO:0016597
PANTHER
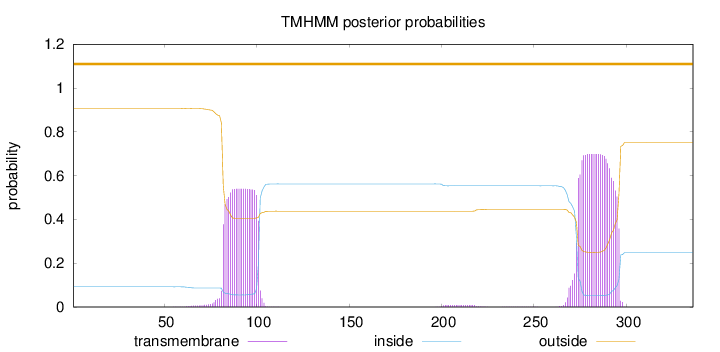
Topology
Length:
336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
26.54036
Exp number, first 60 AAs:
0.00324
Total prob of N-in:
0.09241
outside
1 - 336
Population Genetic Test Statistics
Pi
237.823529
Theta
176.293015
Tajima's D
0.40102
CLR
1.257926
CSRT
0.48307584620769
Interpretation
Uncertain
