Gene
KWMTBOMO05952 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002905
Annotation
acyl-CoA_binding_protein-like_[Bombyx_mori]
Full name
Glycylpeptide N-tetradecanoyltransferase
+ More
Putative acyl-CoA-binding protein
Acyl-CoA-binding protein
Putative acyl-CoA-binding protein
Acyl-CoA-binding protein
Alternative Name
Diazepam-binding inhibitor
Endozepine
Endozepine
Location in the cell
Cytoplasmic Reliability : 1.155 Mitochondrial Reliability : 1.611 Nuclear Reliability : 1.343
Sequence
CDS
ATGTCTCTCGACGAGCAATTCAAACAGGTCGCCGATAAGGTTAGGAACTGGAAGACCAAGCCCAGTGACGATGAGAACCTTGCGCTGTACTCCCTGTACAAGCAGGCTACCATAGGTGATGTTAACATTGCCCAGCCCAGCGGTCTTGTGGAGAGCGCCAAGTGGAAGGCATGGAACGGTCGCAAAGGCATCTCCCAAGACGATGCCAAGAAGCAATACATCGAAAATGCGGAGAAACTCCACTCCAAATACGCATAA
Protein
MSLDEQFKQVADKVRNWKTKPSDDENLALYSLYKQATIGDVNIAQPSGLVESAKWKAWNGRKGISQDDAKKQYIENAEKLHSKYA
Summary
Description
Adds a myristoyl group to the N-terminal glycine residue of certain cellular proteins.
Binds medium- and long-chain acyl-CoA esters with very high affinity and may function as an intracellular carrier of acyl-CoA esters.
Binds medium- and long-chain acyl-CoA esters with very high affinity and may function as an intracellular carrier of acyl-CoA esters.
Catalytic Activity
N-terminal glycyl-[protein] + tetradecanoyl-CoA = CoA + H(+) + N-tetradecanoylglycyl-[protein]
Subunit
Monomer.
Similarity
Belongs to the NMT family.
Belongs to the ACBP family.
Belongs to the ACBP family.
Keywords
Lipid-binding
Transport
Acetylation
Antibiotic
Antimicrobial
Complete proteome
Direct protein sequencing
Endoplasmic reticulum
Golgi apparatus
Phosphoprotein
Reference proteome
Feature
chain Putative acyl-CoA-binding protein
peptide DBI(32-86)
peptide DBI(32-86)
Uniprot
Q9NBL4
B6A8H5
A0A2A4JD00
A0A212EIV0
A0A194QSK1
A0A194Q462
+ More
A0A3S2LW36 A0A0L7LQH8 A0A3B5PQA5 A0A096LVF1 A0A3B3XRJ7 A0A0U1TZF3 A0A3B5MXB6 A0A3B5MR50 A0A3B3TM68 A0A315VKU0 A0A3B3HQS9 H2LZN4 A0A3B3TMA4 A0A3B5MRY1 A0A3P9I9I6 A0A3P9PP87 A0A3P9IA54 L7M062 A0A3B3TKH4 A0A3S3PAZ7 N6TVX9 A0A3P9KQF8 A0A293M2U1 C9W1N3 A0A224YWY9 A0A131Z3T5 A0A3P9KQE8 D6WZ83 A0A3P9PPH4 A0A0L0CCA2 A0A1Z5LGF7 A0A3Q2QPD4 A0A023GEB1 A0A1E1X1P4 G3MN85 A0A3B3V5H0 A0A096LVE3 A0A0P4ZM17 P0C8L7 T1J8I7 A0A2L2XZX7 A0A131XCF6 A0A2I4BTS4 A0A0M3IGG1 A0A3Q2ZNI2 A0A0C9SBL6 A0A1E1XUA6 A0A023FGA8 A0A0P5ESK5 A0A0P5UWT3 D6WZ84 F0J9W9 A0A3P9PP31 E9FZI4 A0A0P4YRG3 J3JW29 A0A087TTJ1 C1LJT2 K7J2N9 A0A023FVY4 A0A0M3JYL8 A0A3B3D5K4 E9GPQ6 A0A151NAC9 I6NMR1 C1LJT1 V8NHT2 A0A3Q2G8T7 A0A3B3BH34 P12026 A0A3Q2C7V2 A0A3Q2QPC3 Q2M0T7 A0A1S3K540 Q6LE27 A0A1U7SME5 A0A1S3K426 E9RJY0 A0A3P7MD27 A0A1D1ZFS7 A0A3B1IXM0 A0A0E9WMX6 A8HG13 A0A1A9ZFV3 H2AX83 A0A1A9V2V4 A0A182G862 A0A232EHZ7 A0A1B0FKR3 A0A087ZSM9 A0A2A3EFW8 A0A3B4E267 A0A084VX07 B4GR37
A0A3S2LW36 A0A0L7LQH8 A0A3B5PQA5 A0A096LVF1 A0A3B3XRJ7 A0A0U1TZF3 A0A3B5MXB6 A0A3B5MR50 A0A3B3TM68 A0A315VKU0 A0A3B3HQS9 H2LZN4 A0A3B3TMA4 A0A3B5MRY1 A0A3P9I9I6 A0A3P9PP87 A0A3P9IA54 L7M062 A0A3B3TKH4 A0A3S3PAZ7 N6TVX9 A0A3P9KQF8 A0A293M2U1 C9W1N3 A0A224YWY9 A0A131Z3T5 A0A3P9KQE8 D6WZ83 A0A3P9PPH4 A0A0L0CCA2 A0A1Z5LGF7 A0A3Q2QPD4 A0A023GEB1 A0A1E1X1P4 G3MN85 A0A3B3V5H0 A0A096LVE3 A0A0P4ZM17 P0C8L7 T1J8I7 A0A2L2XZX7 A0A131XCF6 A0A2I4BTS4 A0A0M3IGG1 A0A3Q2ZNI2 A0A0C9SBL6 A0A1E1XUA6 A0A023FGA8 A0A0P5ESK5 A0A0P5UWT3 D6WZ84 F0J9W9 A0A3P9PP31 E9FZI4 A0A0P4YRG3 J3JW29 A0A087TTJ1 C1LJT2 K7J2N9 A0A023FVY4 A0A0M3JYL8 A0A3B3D5K4 E9GPQ6 A0A151NAC9 I6NMR1 C1LJT1 V8NHT2 A0A3Q2G8T7 A0A3B3BH34 P12026 A0A3Q2C7V2 A0A3Q2QPC3 Q2M0T7 A0A1S3K540 Q6LE27 A0A1U7SME5 A0A1S3K426 E9RJY0 A0A3P7MD27 A0A1D1ZFS7 A0A3B1IXM0 A0A0E9WMX6 A8HG13 A0A1A9ZFV3 H2AX83 A0A1A9V2V4 A0A182G862 A0A232EHZ7 A0A1B0FKR3 A0A087ZSM9 A0A2A3EFW8 A0A3B4E267 A0A084VX07 B4GR37
EC Number
2.3.1.97
Pubmed
11267899
19121390
22118469
26354079
26227816
23542700
+ More
29703783 17554307 25576852 23537049 20650005 24029695 28797301 26830274 18362917 19820115 26108605 28528879 28503490 22216098 26561354 28049606 26131772 29209593 21362191 21292972 22516182 20075255 29451363 22293439 24297900 3289918 8375398 15632085 17994087 26383154 8948484 21382452 25329095 25613341 18519026 22123960 26483478 28648823 24438588
29703783 17554307 25576852 23537049 20650005 24029695 28797301 26830274 18362917 19820115 26108605 28528879 28503490 22216098 26561354 28049606 26131772 29209593 21362191 21292972 22516182 20075255 29451363 22293439 24297900 3289918 8375398 15632085 17994087 26383154 8948484 21382452 25329095 25613341 18519026 22123960 26483478 28648823 24438588
EMBL
BABH01019010
AF246696
AAF78043.1
DQ875235
ABK29477.1
NWSH01002062
+ More
PCG69273.1 AGBW02014561 AGBW02009690 OWR41423.1 OWR50108.1 KQ461175 KPJ07960.1 KQ459472 KPJ00134.1 RSAL01000002 RVE54989.1 JTDY01000324 KOB77702.1 AYCK01006471 AYCK01006472 AYCK01006473 AYCK01006474 AYCK01006475 AYCK01006476 AYCK01006477 EU252315 ACD11893.1 NHOQ01001560 PWA23778.1 GACK01007594 JAA57440.1 NCKU01008582 NCKU01008573 NCKU01000338 NCKU01000337 RWS01809.1 RWS01818.1 RWS15928.1 RWS15936.1 APGK01059191 KB741292 KI207321 ENN70457.1 ERL95657.1 GFWV01010335 MAA35064.1 EZ406185 ACX53980.1 GFPF01007196 MAA18342.1 GEDV01002590 JAP85967.1 KQ971372 EFA10395.1 JRES01000611 KNC29865.1 GFJQ02000732 JAW06238.1 GBBM01003201 GBBM01003200 JAC32217.1 GFAC01006003 JAT93185.1 JO843336 AEO34953.1 GDIP01215053 LRGB01001581 JAJ08349.1 KZS11387.1 CK326514 JH431954 IAAA01006435 LAA01438.1 GEFH01005330 JAP63251.1 GBZX01002897 JAG89843.1 GFAA01000631 JAU02804.1 GBBK01004464 GBBK01004463 JAC20019.1 GDIQ01265097 JAJ86627.1 GDIP01107616 JAL96098.1 EFA10396.1 BK007670 DAA34640.1 GL733192 GL732528 EFX63161.1 EFX87261.1 GDIP01234245 JAI89156.1 APGK01059192 BT127447 AEE62409.1 ENN70458.1 KK116675 KFM68430.1 FN319232 CAX74960.1 GBBL01002408 GBBL01002407 JAC24913.1 UYRR01031289 VDK48604.1 GL732557 EFX78428.1 AKHW03003672 KYO33764.1 JN254628 AEZ53129.1 FN319231 CAX74959.1 AZIM01003864 ETE61511.1 AB019792 DQ885192 CH379069 EAL30839.1 L77976 AAT00460.1 AB618203 BAJ83550.1 UYRU01076779 VDN27344.1 GDJX01002104 JAT65832.1 GBXM01017652 JAH90925.1 EU042104 ABW04128.1 HE650826 CCF58983.1 JXUM01151551 KQ571245 KXJ68217.1 NNAY01004411 OXU17938.1 CCAG010019559 KZ288266 PBC30182.1 ATLV01017829 KE525195 KFB42501.1 CH479188 EDW40222.1
PCG69273.1 AGBW02014561 AGBW02009690 OWR41423.1 OWR50108.1 KQ461175 KPJ07960.1 KQ459472 KPJ00134.1 RSAL01000002 RVE54989.1 JTDY01000324 KOB77702.1 AYCK01006471 AYCK01006472 AYCK01006473 AYCK01006474 AYCK01006475 AYCK01006476 AYCK01006477 EU252315 ACD11893.1 NHOQ01001560 PWA23778.1 GACK01007594 JAA57440.1 NCKU01008582 NCKU01008573 NCKU01000338 NCKU01000337 RWS01809.1 RWS01818.1 RWS15928.1 RWS15936.1 APGK01059191 KB741292 KI207321 ENN70457.1 ERL95657.1 GFWV01010335 MAA35064.1 EZ406185 ACX53980.1 GFPF01007196 MAA18342.1 GEDV01002590 JAP85967.1 KQ971372 EFA10395.1 JRES01000611 KNC29865.1 GFJQ02000732 JAW06238.1 GBBM01003201 GBBM01003200 JAC32217.1 GFAC01006003 JAT93185.1 JO843336 AEO34953.1 GDIP01215053 LRGB01001581 JAJ08349.1 KZS11387.1 CK326514 JH431954 IAAA01006435 LAA01438.1 GEFH01005330 JAP63251.1 GBZX01002897 JAG89843.1 GFAA01000631 JAU02804.1 GBBK01004464 GBBK01004463 JAC20019.1 GDIQ01265097 JAJ86627.1 GDIP01107616 JAL96098.1 EFA10396.1 BK007670 DAA34640.1 GL733192 GL732528 EFX63161.1 EFX87261.1 GDIP01234245 JAI89156.1 APGK01059192 BT127447 AEE62409.1 ENN70458.1 KK116675 KFM68430.1 FN319232 CAX74960.1 GBBL01002408 GBBL01002407 JAC24913.1 UYRR01031289 VDK48604.1 GL732557 EFX78428.1 AKHW03003672 KYO33764.1 JN254628 AEZ53129.1 FN319231 CAX74959.1 AZIM01003864 ETE61511.1 AB019792 DQ885192 CH379069 EAL30839.1 L77976 AAT00460.1 AB618203 BAJ83550.1 UYRU01076779 VDN27344.1 GDJX01002104 JAT65832.1 GBXM01017652 JAH90925.1 EU042104 ABW04128.1 HE650826 CCF58983.1 JXUM01151551 KQ571245 KXJ68217.1 NNAY01004411 OXU17938.1 CCAG010019559 KZ288266 PBC30182.1 ATLV01017829 KE525195 KFB42501.1 CH479188 EDW40222.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000002852 UP000028760 UP000261480 UP000261380 UP000261500 UP000001038 UP000265200 UP000242638 UP000285301 UP000019118 UP000030742 UP000265180 UP000007266 UP000037069 UP000265000 UP000076858 UP000192220 UP000036681 UP000264800 UP000000305 UP000054359 UP000002358 UP000036680 UP000261560 UP000050525 UP000265020 UP000008227 UP000001819 UP000085678 UP000189705 UP000018467 UP000092445 UP000005220 UP000078200 UP000069940 UP000249989 UP000215335 UP000092444 UP000005203 UP000242457 UP000261440 UP000030765 UP000008744
UP000037510 UP000002852 UP000028760 UP000261480 UP000261380 UP000261500 UP000001038 UP000265200 UP000242638 UP000285301 UP000019118 UP000030742 UP000265180 UP000007266 UP000037069 UP000265000 UP000076858 UP000192220 UP000036681 UP000264800 UP000000305 UP000054359 UP000002358 UP000036680 UP000261560 UP000050525 UP000265020 UP000008227 UP000001819 UP000085678 UP000189705 UP000018467 UP000092445 UP000005220 UP000078200 UP000069940 UP000249989 UP000215335 UP000092444 UP000005203 UP000242457 UP000261440 UP000030765 UP000008744
Interpro
IPR014352
FERM/acyl-CoA-bd_prot_sf
+ More
IPR035984 Acyl-CoA-binding_sf
IPR000582 Acyl-CoA-binding_protein
IPR022408 Acyl-CoA-binding_prot_CS
IPR022677 MyristoylCoA_TrFase_C
IPR000903 MyristoylCoA_TrFase
IPR022676 MyristoylCoA_TrFase_N
IPR022678 MyristoylCoA_TrFase_CS
IPR016181 Acyl_CoA_acyltransferase
IPR035984 Acyl-CoA-binding_sf
IPR000582 Acyl-CoA-binding_protein
IPR022408 Acyl-CoA-binding_prot_CS
IPR022677 MyristoylCoA_TrFase_C
IPR000903 MyristoylCoA_TrFase
IPR022676 MyristoylCoA_TrFase_N
IPR022678 MyristoylCoA_TrFase_CS
IPR016181 Acyl_CoA_acyltransferase
Gene 3D
CDD
ProteinModelPortal
Q9NBL4
B6A8H5
A0A2A4JD00
A0A212EIV0
A0A194QSK1
A0A194Q462
+ More
A0A3S2LW36 A0A0L7LQH8 A0A3B5PQA5 A0A096LVF1 A0A3B3XRJ7 A0A0U1TZF3 A0A3B5MXB6 A0A3B5MR50 A0A3B3TM68 A0A315VKU0 A0A3B3HQS9 H2LZN4 A0A3B3TMA4 A0A3B5MRY1 A0A3P9I9I6 A0A3P9PP87 A0A3P9IA54 L7M062 A0A3B3TKH4 A0A3S3PAZ7 N6TVX9 A0A3P9KQF8 A0A293M2U1 C9W1N3 A0A224YWY9 A0A131Z3T5 A0A3P9KQE8 D6WZ83 A0A3P9PPH4 A0A0L0CCA2 A0A1Z5LGF7 A0A3Q2QPD4 A0A023GEB1 A0A1E1X1P4 G3MN85 A0A3B3V5H0 A0A096LVE3 A0A0P4ZM17 P0C8L7 T1J8I7 A0A2L2XZX7 A0A131XCF6 A0A2I4BTS4 A0A0M3IGG1 A0A3Q2ZNI2 A0A0C9SBL6 A0A1E1XUA6 A0A023FGA8 A0A0P5ESK5 A0A0P5UWT3 D6WZ84 F0J9W9 A0A3P9PP31 E9FZI4 A0A0P4YRG3 J3JW29 A0A087TTJ1 C1LJT2 K7J2N9 A0A023FVY4 A0A0M3JYL8 A0A3B3D5K4 E9GPQ6 A0A151NAC9 I6NMR1 C1LJT1 V8NHT2 A0A3Q2G8T7 A0A3B3BH34 P12026 A0A3Q2C7V2 A0A3Q2QPC3 Q2M0T7 A0A1S3K540 Q6LE27 A0A1U7SME5 A0A1S3K426 E9RJY0 A0A3P7MD27 A0A1D1ZFS7 A0A3B1IXM0 A0A0E9WMX6 A8HG13 A0A1A9ZFV3 H2AX83 A0A1A9V2V4 A0A182G862 A0A232EHZ7 A0A1B0FKR3 A0A087ZSM9 A0A2A3EFW8 A0A3B4E267 A0A084VX07 B4GR37
A0A3S2LW36 A0A0L7LQH8 A0A3B5PQA5 A0A096LVF1 A0A3B3XRJ7 A0A0U1TZF3 A0A3B5MXB6 A0A3B5MR50 A0A3B3TM68 A0A315VKU0 A0A3B3HQS9 H2LZN4 A0A3B3TMA4 A0A3B5MRY1 A0A3P9I9I6 A0A3P9PP87 A0A3P9IA54 L7M062 A0A3B3TKH4 A0A3S3PAZ7 N6TVX9 A0A3P9KQF8 A0A293M2U1 C9W1N3 A0A224YWY9 A0A131Z3T5 A0A3P9KQE8 D6WZ83 A0A3P9PPH4 A0A0L0CCA2 A0A1Z5LGF7 A0A3Q2QPD4 A0A023GEB1 A0A1E1X1P4 G3MN85 A0A3B3V5H0 A0A096LVE3 A0A0P4ZM17 P0C8L7 T1J8I7 A0A2L2XZX7 A0A131XCF6 A0A2I4BTS4 A0A0M3IGG1 A0A3Q2ZNI2 A0A0C9SBL6 A0A1E1XUA6 A0A023FGA8 A0A0P5ESK5 A0A0P5UWT3 D6WZ84 F0J9W9 A0A3P9PP31 E9FZI4 A0A0P4YRG3 J3JW29 A0A087TTJ1 C1LJT2 K7J2N9 A0A023FVY4 A0A0M3JYL8 A0A3B3D5K4 E9GPQ6 A0A151NAC9 I6NMR1 C1LJT1 V8NHT2 A0A3Q2G8T7 A0A3B3BH34 P12026 A0A3Q2C7V2 A0A3Q2QPC3 Q2M0T7 A0A1S3K540 Q6LE27 A0A1U7SME5 A0A1S3K426 E9RJY0 A0A3P7MD27 A0A1D1ZFS7 A0A3B1IXM0 A0A0E9WMX6 A8HG13 A0A1A9ZFV3 H2AX83 A0A1A9V2V4 A0A182G862 A0A232EHZ7 A0A1B0FKR3 A0A087ZSM9 A0A2A3EFW8 A0A3B4E267 A0A084VX07 B4GR37
PDB
2CB8
E-value=3.39006e-13,
Score=175
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum
Golgi localization is dependent on ligand binding. With evidence from 2 publications.
Golgi apparatus Golgi localization is dependent on ligand binding. With evidence from 2 publications.
Golgi apparatus Golgi localization is dependent on ligand binding. With evidence from 2 publications.
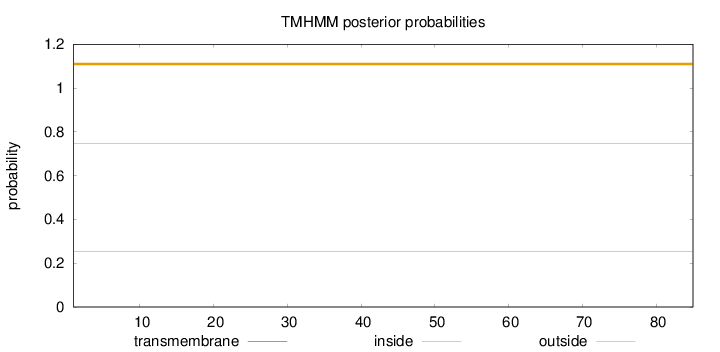
Length:
85
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00103
Exp number, first 60 AAs:
0.00103
Total prob of N-in:
0.25288
outside
1 - 85
Population Genetic Test Statistics
Pi
268.208361
Theta
192.763266
Tajima's D
1.183237
CLR
0
CSRT
0.706864656767162
Interpretation
Uncertain
