Gene
KWMTBOMO05948 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002907
Annotation
diapause_bioclock_protein_[Bombyx_mori]
Full name
Superoxide dismutase [Cu-Zn]
Location in the cell
Extracellular Reliability : 2.648
Sequence
CDS
ATGCTGCTTCAACTCACATTCCTGGCTGCGATCGCTCTGGCGACGGCTCATCATGGCTTCACCACGCCGTCCCGCGCCATTGCCTTTCTATCCACAGAAACGATTAGAGGCAACATCACATTCACTCAGGTTCAAGATGGGAAGGTCCACGTTCAGGGGGGCATCACTGGGCTGCCTCCGGGTGAATACGGCTTCCACGTCCATGAGAAGGGCGACCTAAGCGGCGGTTGTGTATCGACTGGATCTCATTTCAATCCTGAACATAAGGACCACGGTCATCCGAACGATGTTAACCGTCACGTCGGCGACCTTGGAAACGTGGTCTTTGACGAGAACCACTACAGCAGGATCGACCTGGTAGACGACCAGATCTCGCTATCGGGTCCGCACGGTATCATCGGCAGGGCGGTGGTGCTCCACGAGAAAGCTGATGATTACGGCAAGAGTGACCATCCGGACTCCAGGAAGACCGGCAACGCTGGCGGTCGAGTCGCTTGCGGTGTCATCGGAATTCTATAG
Protein
MLLQLTFLAAIALATAHHGFTTPSRAIAFLSTETIRGNITFTQVQDGKVHVQGGITGLPPGEYGFHVHEKGDLSGGCVSTGSHFNPEHKDHGHPNDVNRHVGDLGNVVFDENHYSRIDLVDDQISLSGPHGIIGRAVVLHEKADDYGKSDHPDSRKTGNAGGRVACGVIGIL
Summary
Description
Destroys radicals which are normally produced within the cells and which are toxic to biological systems.
Catalytic Activity
2 H(+) + 2 superoxide = H2O2 + O2
Cofactor
Cu cation
Zn(2+)
Zn(2+)
Biophysicochemical Properties
Active between pH 5 and 10.
Subunit
Homodimer (Probable).
Similarity
Belongs to the Cu-Zn superoxide dismutase family.
Keywords
Antioxidant
Copper
Cytoplasm
Direct protein sequencing
Metal-binding
Oxidoreductase
Zinc
Feature
chain Superoxide dismutase [Cu-Zn]
Uniprot
Q4JHW1
Q3Y666
Q08J22
H9J072
H9BE67
I4DMD1
+ More
A0A194QWK9 A0A0L7L5I0 A0A194Q551 A0A0K0KJ13 A0A2S0RQQ0 A0A2H4G7I9 A0A3S2MA90 A0A212F790 A0A3S5HJH3 A0A2H1W8P0 A0A194QSJ6 A0A2A4JF55 A0A194Q3L2 A0A088SGB6 A0A034WIM4 A0A0C9REV0 A0A034WL86 A0A0C9R2T7 A0A0K8U5U3 A0A0K8UZT3 A0A0A1XL41 M1J3Y8 A0A0A1XSI9 A0A2P5YCL6 A0A1U8IBS9 A0A0B0MPF5 A0A0D2TZK0 A0A182NQ77 A0A034WMM4 T1E7D6 A0A1Y9H174 A0A1Y9H7N9 A0A1Q3B854 E6Y8N8 A8CWF4 A0A2P5Q434 A0A1U8IBW9 A0A1B0CYC5 A0A1U7ZG86 A0A2M3ZD70 A0A2S3H514 A0A2T7F042 A0A1J1J1H2 A0A182QE62 A0A2M4AYQ5 A0A1I8JVG9 A0A1Y9IQV8 A0A182LGP2 A0A182UJ15 A7UTA9 A0A182HNW0 K3ZXQ6 A0A1Q3F7B4 A0A1J1IWP5 A0A240PME8 A0A182FCV9 A0A087YZB1 H6S1K6 A0A182WME1 W5JT45 A0A2M3ZDC7 A0A182J3Z1 A0A1S6PDB1 A0A1I8JSG9 A0A2M4BZQ2 A0A182P8U8 A0A1J1IYC6 C6K2H2 A0A182XET3 A0A125SL96 A0A182YC20 A0A1C7CZ12 A0A1I8JSX4 A0A182VG23 Q7Q9H5 L7S334 A0A2I4G8M7 A0A1I8JUZ9 A0A2C9GUF8 A0A1Y9IV76 A0A2M4BZW3 A9LNG3 C0HK70 Q6QVQ5 A9LNF7 A0A182RV04 A9LNH0 Q5EGZ2 A9LMH9 H9A9E0 A0A3F2YTW1 Q71S31 A0A182MMA7
A0A194QWK9 A0A0L7L5I0 A0A194Q551 A0A0K0KJ13 A0A2S0RQQ0 A0A2H4G7I9 A0A3S2MA90 A0A212F790 A0A3S5HJH3 A0A2H1W8P0 A0A194QSJ6 A0A2A4JF55 A0A194Q3L2 A0A088SGB6 A0A034WIM4 A0A0C9REV0 A0A034WL86 A0A0C9R2T7 A0A0K8U5U3 A0A0K8UZT3 A0A0A1XL41 M1J3Y8 A0A0A1XSI9 A0A2P5YCL6 A0A1U8IBS9 A0A0B0MPF5 A0A0D2TZK0 A0A182NQ77 A0A034WMM4 T1E7D6 A0A1Y9H174 A0A1Y9H7N9 A0A1Q3B854 E6Y8N8 A8CWF4 A0A2P5Q434 A0A1U8IBW9 A0A1B0CYC5 A0A1U7ZG86 A0A2M3ZD70 A0A2S3H514 A0A2T7F042 A0A1J1J1H2 A0A182QE62 A0A2M4AYQ5 A0A1I8JVG9 A0A1Y9IQV8 A0A182LGP2 A0A182UJ15 A7UTA9 A0A182HNW0 K3ZXQ6 A0A1Q3F7B4 A0A1J1IWP5 A0A240PME8 A0A182FCV9 A0A087YZB1 H6S1K6 A0A182WME1 W5JT45 A0A2M3ZDC7 A0A182J3Z1 A0A1S6PDB1 A0A1I8JSG9 A0A2M4BZQ2 A0A182P8U8 A0A1J1IYC6 C6K2H2 A0A182XET3 A0A125SL96 A0A182YC20 A0A1C7CZ12 A0A1I8JSX4 A0A182VG23 Q7Q9H5 L7S334 A0A2I4G8M7 A0A1I8JUZ9 A0A2C9GUF8 A0A1Y9IV76 A0A2M4BZW3 A9LNG3 C0HK70 Q6QVQ5 A9LNF7 A0A182RV04 A9LNH0 Q5EGZ2 A9LMH9 H9A9E0 A0A3F2YTW1 Q71S31 A0A182MMA7
EC Number
1.15.1.1
Pubmed
EMBL
DQ086424
EF121772
EF121773
AAY86076.1
ABL63513.1
ABL63514.2
+ More
DQ154907 AAZ80044.1 AB257197 LC424927 BAF34334.1 BBG40623.1 BABH01019014 JQ003185 AFC35302.1 AK402449 BAM19071.1 KQ461175 KPJ07956.1 JTDY01002860 KOB70586.1 KQ459472 KPJ00130.1 KM215270 AIJ00867.1 MF797882 AWA45436.1 KU356855 APW79687.1 RSAL01000002 RVE54993.1 AGBW02009903 OWR49600.1 MH374164 AZK16058.1 ODYU01006924 SOQ49202.1 KPJ07955.1 NWSH01001766 PCG70204.1 KPJ00128.1 KM384740 AIO11231.1 GAKP01003531 JAC55421.1 GBYB01006885 JAG76652.1 GAKP01003533 JAC55419.1 GBYB01001151 JAG70918.1 GDHF01030393 GDHF01027182 GDHF01019979 GDHF01018890 GDHF01015981 GDHF01011959 JAI21921.1 JAI25132.1 JAI32335.1 JAI33424.1 JAI36333.1 JAI40355.1 GDHF01020198 JAI32116.1 GBXI01002213 JAD12079.1 JX263432 AGE89778.1 GBXI01000467 JAD13825.1 KZ663361 PPS13340.1 JRRC01403715 KHG04013.1 CM001752 KJB80823.1 GAKP01003532 JAC55420.1 GAMD01003137 JAA98453.1 AXCN02001018 BDDD01000337 GAV64207.1 FJ589638 ACU52586.1 EU124625 ABV60343.1 KZ650042 PPD69244.1 AJWK01010399 AJWK01010400 AJWK01010401 AJWK01010402 AJWK01010403 GGFM01005701 MBW26452.1 CM008047 PAN15435.1 CM009750 PUZ73440.1 CVRI01000063 CRL04705.1 GGFK01012593 MBW45914.1 AAAB01008900 EDO64116.1 APCN01002794 AGNK02001362 CM003529 RCV14396.1 GFDL01011585 JAV23460.1 CRL04707.1 GU731670 ADN04915.1 ADMH02000552 ETN65899.1 GGFM01005727 MBW26478.1 KX925930 AQU42894.1 GGFJ01009404 MBW58545.1 CRL04706.1 GQ149102 ACS45202.1 KT594770 AMH40758.1 EAA09396.4 JX948790 AGC13159.1 AXCM01008451 GGFJ01009403 MBW58544.1 EU250773 ABX54855.1 AY524130 AAS17758.1 EU250767 ABX54849.1 EU250780 ABX54862.1 AY881165 AAW80441.1 EU247081 ABX26134.1 JN869247 AFC35179.1 AF318938 AAQ14591.1
DQ154907 AAZ80044.1 AB257197 LC424927 BAF34334.1 BBG40623.1 BABH01019014 JQ003185 AFC35302.1 AK402449 BAM19071.1 KQ461175 KPJ07956.1 JTDY01002860 KOB70586.1 KQ459472 KPJ00130.1 KM215270 AIJ00867.1 MF797882 AWA45436.1 KU356855 APW79687.1 RSAL01000002 RVE54993.1 AGBW02009903 OWR49600.1 MH374164 AZK16058.1 ODYU01006924 SOQ49202.1 KPJ07955.1 NWSH01001766 PCG70204.1 KPJ00128.1 KM384740 AIO11231.1 GAKP01003531 JAC55421.1 GBYB01006885 JAG76652.1 GAKP01003533 JAC55419.1 GBYB01001151 JAG70918.1 GDHF01030393 GDHF01027182 GDHF01019979 GDHF01018890 GDHF01015981 GDHF01011959 JAI21921.1 JAI25132.1 JAI32335.1 JAI33424.1 JAI36333.1 JAI40355.1 GDHF01020198 JAI32116.1 GBXI01002213 JAD12079.1 JX263432 AGE89778.1 GBXI01000467 JAD13825.1 KZ663361 PPS13340.1 JRRC01403715 KHG04013.1 CM001752 KJB80823.1 GAKP01003532 JAC55420.1 GAMD01003137 JAA98453.1 AXCN02001018 BDDD01000337 GAV64207.1 FJ589638 ACU52586.1 EU124625 ABV60343.1 KZ650042 PPD69244.1 AJWK01010399 AJWK01010400 AJWK01010401 AJWK01010402 AJWK01010403 GGFM01005701 MBW26452.1 CM008047 PAN15435.1 CM009750 PUZ73440.1 CVRI01000063 CRL04705.1 GGFK01012593 MBW45914.1 AAAB01008900 EDO64116.1 APCN01002794 AGNK02001362 CM003529 RCV14396.1 GFDL01011585 JAV23460.1 CRL04707.1 GU731670 ADN04915.1 ADMH02000552 ETN65899.1 GGFM01005727 MBW26478.1 KX925930 AQU42894.1 GGFJ01009404 MBW58545.1 CRL04706.1 GQ149102 ACS45202.1 KT594770 AMH40758.1 EAA09396.4 JX948790 AGC13159.1 AXCM01008451 GGFJ01009403 MBW58544.1 EU250773 ABX54855.1 AY524130 AAS17758.1 EU250767 ABX54849.1 EU250780 ABX54862.1 AY881165 AAW80441.1 EU247081 ABX26134.1 JN869247 AFC35179.1 AF318938 AAQ14591.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000283053
UP000007151
+ More
UP000218220 UP000239757 UP000189702 UP000032304 UP000075884 UP000075886 UP000187406 UP000239050 UP000092461 UP000189703 UP000243499 UP000244336 UP000183832 UP000076407 UP000075903 UP000075882 UP000075902 UP000007062 UP000075840 UP000004995 UP000075880 UP000069272 UP000075920 UP000000673 UP000075885 UP000076408 UP000235220 UP000075883 UP000075900 UP000075881
UP000218220 UP000239757 UP000189702 UP000032304 UP000075884 UP000075886 UP000187406 UP000239050 UP000092461 UP000189703 UP000243499 UP000244336 UP000183832 UP000076407 UP000075903 UP000075882 UP000075902 UP000007062 UP000075840 UP000004995 UP000075880 UP000069272 UP000075920 UP000000673 UP000075885 UP000076408 UP000235220 UP000075883 UP000075900 UP000075881
Interpro
SUPFAM
SSF49329
SSF49329
Gene 3D
ProteinModelPortal
Q4JHW1
Q3Y666
Q08J22
H9J072
H9BE67
I4DMD1
+ More
A0A194QWK9 A0A0L7L5I0 A0A194Q551 A0A0K0KJ13 A0A2S0RQQ0 A0A2H4G7I9 A0A3S2MA90 A0A212F790 A0A3S5HJH3 A0A2H1W8P0 A0A194QSJ6 A0A2A4JF55 A0A194Q3L2 A0A088SGB6 A0A034WIM4 A0A0C9REV0 A0A034WL86 A0A0C9R2T7 A0A0K8U5U3 A0A0K8UZT3 A0A0A1XL41 M1J3Y8 A0A0A1XSI9 A0A2P5YCL6 A0A1U8IBS9 A0A0B0MPF5 A0A0D2TZK0 A0A182NQ77 A0A034WMM4 T1E7D6 A0A1Y9H174 A0A1Y9H7N9 A0A1Q3B854 E6Y8N8 A8CWF4 A0A2P5Q434 A0A1U8IBW9 A0A1B0CYC5 A0A1U7ZG86 A0A2M3ZD70 A0A2S3H514 A0A2T7F042 A0A1J1J1H2 A0A182QE62 A0A2M4AYQ5 A0A1I8JVG9 A0A1Y9IQV8 A0A182LGP2 A0A182UJ15 A7UTA9 A0A182HNW0 K3ZXQ6 A0A1Q3F7B4 A0A1J1IWP5 A0A240PME8 A0A182FCV9 A0A087YZB1 H6S1K6 A0A182WME1 W5JT45 A0A2M3ZDC7 A0A182J3Z1 A0A1S6PDB1 A0A1I8JSG9 A0A2M4BZQ2 A0A182P8U8 A0A1J1IYC6 C6K2H2 A0A182XET3 A0A125SL96 A0A182YC20 A0A1C7CZ12 A0A1I8JSX4 A0A182VG23 Q7Q9H5 L7S334 A0A2I4G8M7 A0A1I8JUZ9 A0A2C9GUF8 A0A1Y9IV76 A0A2M4BZW3 A9LNG3 C0HK70 Q6QVQ5 A9LNF7 A0A182RV04 A9LNH0 Q5EGZ2 A9LMH9 H9A9E0 A0A3F2YTW1 Q71S31 A0A182MMA7
A0A194QWK9 A0A0L7L5I0 A0A194Q551 A0A0K0KJ13 A0A2S0RQQ0 A0A2H4G7I9 A0A3S2MA90 A0A212F790 A0A3S5HJH3 A0A2H1W8P0 A0A194QSJ6 A0A2A4JF55 A0A194Q3L2 A0A088SGB6 A0A034WIM4 A0A0C9REV0 A0A034WL86 A0A0C9R2T7 A0A0K8U5U3 A0A0K8UZT3 A0A0A1XL41 M1J3Y8 A0A0A1XSI9 A0A2P5YCL6 A0A1U8IBS9 A0A0B0MPF5 A0A0D2TZK0 A0A182NQ77 A0A034WMM4 T1E7D6 A0A1Y9H174 A0A1Y9H7N9 A0A1Q3B854 E6Y8N8 A8CWF4 A0A2P5Q434 A0A1U8IBW9 A0A1B0CYC5 A0A1U7ZG86 A0A2M3ZD70 A0A2S3H514 A0A2T7F042 A0A1J1J1H2 A0A182QE62 A0A2M4AYQ5 A0A1I8JVG9 A0A1Y9IQV8 A0A182LGP2 A0A182UJ15 A7UTA9 A0A182HNW0 K3ZXQ6 A0A1Q3F7B4 A0A1J1IWP5 A0A240PME8 A0A182FCV9 A0A087YZB1 H6S1K6 A0A182WME1 W5JT45 A0A2M3ZDC7 A0A182J3Z1 A0A1S6PDB1 A0A1I8JSG9 A0A2M4BZQ2 A0A182P8U8 A0A1J1IYC6 C6K2H2 A0A182XET3 A0A125SL96 A0A182YC20 A0A1C7CZ12 A0A1I8JSX4 A0A182VG23 Q7Q9H5 L7S334 A0A2I4G8M7 A0A1I8JUZ9 A0A2C9GUF8 A0A1Y9IV76 A0A2M4BZW3 A9LNG3 C0HK70 Q6QVQ5 A9LNF7 A0A182RV04 A9LNH0 Q5EGZ2 A9LMH9 H9A9E0 A0A3F2YTW1 Q71S31 A0A182MMA7
PDB
2E46
E-value=5.88602e-75,
Score=709
Ontologies
PATHWAY
GO
PANTHER
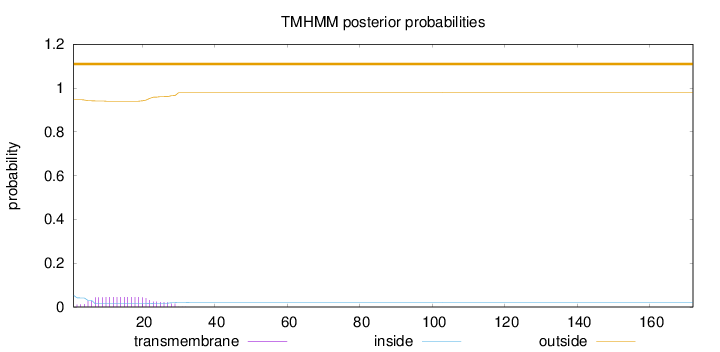
Topology
Subcellular location
Cytoplasm
SignalP
Position: 1 - 16,
Likelihood: 0.962064
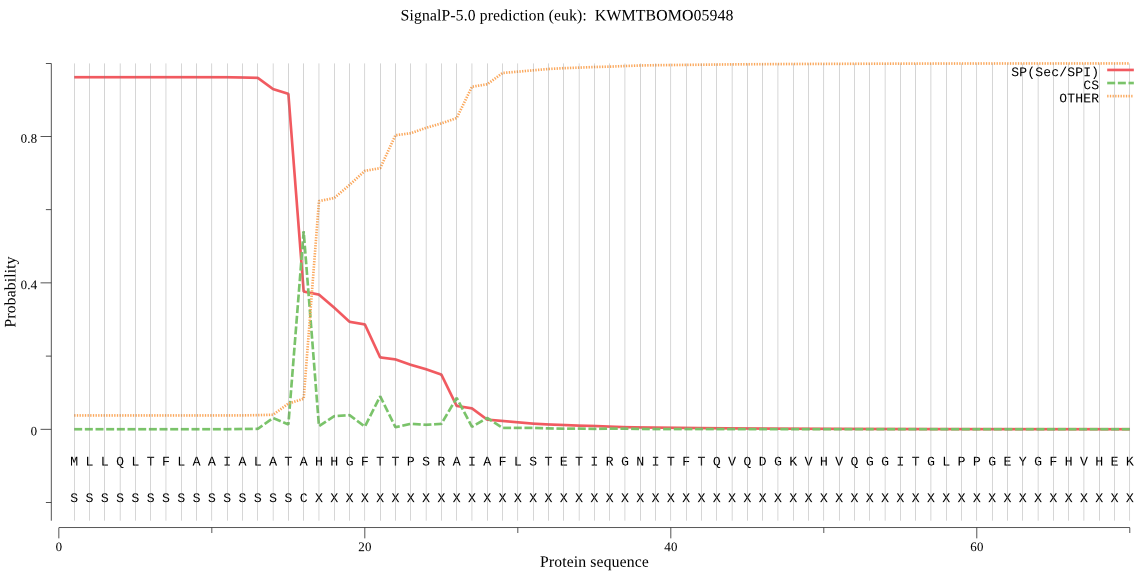
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.942139999999999
Exp number, first 60 AAs:
0.94197
Total prob of N-in:
0.05313
outside
1 - 172
Population Genetic Test Statistics
Pi
389.713984
Theta
193.104443
Tajima's D
2.694206
CLR
1.050301
CSRT
0.961101944902755
Interpretation
Uncertain
