Gene
KWMTBOMO05946
Annotation
PREDICTED:_23_kDa_integral_membrane_protein-like_[Bombyx_mori]
Full name
Tetraspanin
+ More
Tetraspanin-9
Tetraspanin-9
Location in the cell
PlasmaMembrane Reliability : 4.143
Sequence
CDS
ATGAAACCCCGCTCTATACGCTGCATGAAGTTTTTTCTGACGCTCTTCAATGTGATTTGCATCGTCTTTGGTTTGATTTTGATGGCCATTTGCGTCATTAATATGCGCGACAAGAAGAATCGTCCGGATCAACAATCAGCTTTGTCAAGAGGTGTGCTGTCATTCCTCTTAACTCTTGGTTTCTGTTTGGCTGTAACTGGTGTTTTGGGCTGCGTCGGTGCTTTGAGAGAGCACGTTAAAGTACTTTATGTGCATGCTTGCTTCTTCATATTCCTGGTATCAGTCGAGCTGGTGGTAGGAGTCGGAGGGGCGGTCCTTAGCGCTTGGGTCGGCGGGAGCAACGAACTACGAATGCAATTTTACAAAAACGCTACCGTGGAAGACGAAGTATCGAAACATCACAATTTCTGGGACAGCTTACAGTCCGAGAATAAATGTTGTGGCGTGGACGGTCCCCAAGATTACGCCATACTGCATCGAGACATTCCGGCCTCATGCTGCGCCCGAGCTTATCCTCTCCGCGAGGGTGGAGCCCGTCGACATTTGTACGCTACCTGTATTTCGGAAAGGACATACTACAAGGCAGGTTGCGAAGAGGCTCTCCGACAGAAGAAAACCATGAAAGGAAACATTTTTATTAGTACAGGAATCGTTTTCGCTTTATTGGAGGTACGTGTAAAAATATTTTTAGTTACAAAATTACATGATCATTTTTATAATATAGACTAG
Protein
MKPRSIRCMKFFLTLFNVICIVFGLILMAICVINMRDKKNRPDQQSALSRGVLSFLLTLGFCLAVTGVLGCVGALREHVKVLYVHACFFIFLVSVELVVGVGGAVLSAWVGGSNELRMQFYKNATVEDEVSKHHNFWDSLQSENKCCGVDGPQDYAILHRDIPASCCARAYPLREGGARRHLYATCISERTYYKAGCEEALRQKKTMKGNIFISTGIVFALLEVRVKIFLVTKLHDHFYNID
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Tetraspanin-9
Uniprot
A0A2A4JFK4
A0A2H1W865
A0A212F783
A0A0L7KZ26
A0A194QRJ2
A0A194Q3L9
+ More
A0A212F781 A0A194Q457 A0A3S2P1W8 A0A3S2LJ58 A0A2A4JFM4 A0A212F793 A0A194Q9P1 A0A2H8TQA1 A0A2H1W9Q9 A0A087TFG6 A0A076KUR5 K7J9H7 A0A0B5H7U5 A0A182FCW0 A0A0K8WBF2 N6UM92 A0A0A1WG71 A0A084W472 A0A034WD08 A0A2S2QQ20 A0A182RV02 A0A1B0D9D5 B4KR28 B4MQJ7 Q7Q9H6 A0A182XET4 A0A182HNW1 T1GNT4 A0A232EUB7 A0A0C9QE23 Q28Y97 A0A1B0CUN9 A0A182MBF2 A0A182WME0 B4LQ51 H7CHJ1 B4J8Z2 A0A026W8X0 A0A182P8U7 A0A0J7L311 W8BXV6 B4GCW3 A0A1D1UQ65 Q176T5 A0A0M5J749 A0A026W8U6 A0A3B5KLI5 A0A0V0JAW3 A0A158NAZ0 A0A3B3RMI6 A0A182YC21 A0A2U9BBA8 A0A0N0BFU5 F6TTN6 N6UQ61 A0A3B0JKU2 A0A0N0BD54 A0A1Z5L5C1 H2UYH5 A0A3Q1FFI0 D6X0S1 A0A3B3RJZ9 A0A0C9RAA1 W5JSN2 A0A336MEI8 E2BYV0 A0A3B3ULN6 H2YU34 A0A1I8QDE5 K7ISS9 A0A1W4XT51 A0A1I8MII5 A0A151HY78 D6X0S0 A0A3P8YU62 Q6GMK6 A0A3B3RM52 A0A3Q2ZL31 B3MFB3 A1Z8K7 A0A0L7KTN2 E2B224
A0A212F781 A0A194Q457 A0A3S2P1W8 A0A3S2LJ58 A0A2A4JFM4 A0A212F793 A0A194Q9P1 A0A2H8TQA1 A0A2H1W9Q9 A0A087TFG6 A0A076KUR5 K7J9H7 A0A0B5H7U5 A0A182FCW0 A0A0K8WBF2 N6UM92 A0A0A1WG71 A0A084W472 A0A034WD08 A0A2S2QQ20 A0A182RV02 A0A1B0D9D5 B4KR28 B4MQJ7 Q7Q9H6 A0A182XET4 A0A182HNW1 T1GNT4 A0A232EUB7 A0A0C9QE23 Q28Y97 A0A1B0CUN9 A0A182MBF2 A0A182WME0 B4LQ51 H7CHJ1 B4J8Z2 A0A026W8X0 A0A182P8U7 A0A0J7L311 W8BXV6 B4GCW3 A0A1D1UQ65 Q176T5 A0A0M5J749 A0A026W8U6 A0A3B5KLI5 A0A0V0JAW3 A0A158NAZ0 A0A3B3RMI6 A0A182YC21 A0A2U9BBA8 A0A0N0BFU5 F6TTN6 N6UQ61 A0A3B0JKU2 A0A0N0BD54 A0A1Z5L5C1 H2UYH5 A0A3Q1FFI0 D6X0S1 A0A3B3RJZ9 A0A0C9RAA1 W5JSN2 A0A336MEI8 E2BYV0 A0A3B3ULN6 H2YU34 A0A1I8QDE5 K7ISS9 A0A1W4XT51 A0A1I8MII5 A0A151HY78 D6X0S0 A0A3P8YU62 Q6GMK6 A0A3B3RM52 A0A3Q2ZL31 B3MFB3 A1Z8K7 A0A0L7KTN2 E2B224
Pubmed
22118469
26227816
26354079
20075255
23537049
25830018
+ More
24438588 25348373 17994087 12364791 28648823 15632085 24508170 30249741 24495485 27649274 17510324 21551351 21347285 29240929 25244985 12481130 15114417 28528879 18362917 19820115 20920257 23761445 20798317 25315136 25069045 23594743 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
24438588 25348373 17994087 12364791 28648823 15632085 24508170 30249741 24495485 27649274 17510324 21551351 21347285 29240929 25244985 12481130 15114417 28528879 18362917 19820115 20920257 23761445 20798317 25315136 25069045 23594743 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
NWSH01001766
PCG70203.1
ODYU01006924
SOQ49203.1
AGBW02009903
OWR49602.1
+ More
JTDY01004293 KOB68321.1 KQ461175 KPJ07954.1 KQ459472 KPJ00127.1 OWR49601.1 KPJ00129.1 RSAL01000002 RVE54994.1 RVE54996.1 PCG70202.1 OWR49603.1 KPJ00126.1 GFXV01004355 MBW16160.1 SOQ49204.1 KK114976 KFM63855.1 KF433416 AII97740.1 KJ535125 AJF38198.1 GDHF01004139 JAI48175.1 APGK01026523 APGK01026524 KB740635 KB631992 ENN79832.1 ERL87761.1 GBXI01016899 JAC97392.1 ATLV01020284 KE525297 KFB45016.1 GAKP01006771 GAKP01006770 JAC52182.1 GGMS01010625 MBY79828.1 AJVK01012925 CH933808 EDW10384.2 CH963849 EDW74386.1 AAAB01008900 EAA09395.4 APCN01002794 CAQQ02119555 CAQQ02119556 CAQQ02119557 CAQQ02119558 CAQQ02119559 CAQQ02119560 NNAY01002144 OXU21942.1 GBYB01012658 JAG82425.1 CM000071 EAL26068.1 AJWK01029481 AXCM01011569 CH940648 EDW61336.1 AB701637 BAL70289.1 CH916367 EDW01341.1 KK107353 QOIP01000012 EZA52081.1 RLU15762.1 LBMM01001031 KMQ97046.1 GAMC01002393 JAC04163.1 CH479181 EDW32526.1 BDGG01000001 GAU88503.1 CH477383 EAT42156.1 CP012524 ALC41286.1 KK107324 QOIP01000006 EZA52517.1 RLU21332.1 GEEE01000265 JAP62960.1 ADTU01010579 ADTU01010580 ADTU01010581 CP026247 AWP01225.1 KQ435794 KOX73598.1 EAAA01000909 APGK01008978 APGK01008979 APGK01008980 APGK01008981 KB737825 ENN82906.1 OUUW01000001 SPP73856.1 KQ435878 KOX69968.1 GFJQ02004357 JAW02613.1 KQ971372 EFA09563.1 GBYB01003736 JAG73503.1 ADMH02000552 ETN65900.1 UFQS01000997 UFQT01000997 SSX08337.1 SSX28360.1 GL451531 EFN79122.1 EFN79123.1 AAZX01003292 KQ976724 KYM76647.1 EFA09564.2 BX901962 CT573114 BC074038 BC142792 CH902619 EDV35587.1 AE013599 AAM68735.1 JTDY01005790 KOB66627.1 GL445031 EFN60280.1
JTDY01004293 KOB68321.1 KQ461175 KPJ07954.1 KQ459472 KPJ00127.1 OWR49601.1 KPJ00129.1 RSAL01000002 RVE54994.1 RVE54996.1 PCG70202.1 OWR49603.1 KPJ00126.1 GFXV01004355 MBW16160.1 SOQ49204.1 KK114976 KFM63855.1 KF433416 AII97740.1 KJ535125 AJF38198.1 GDHF01004139 JAI48175.1 APGK01026523 APGK01026524 KB740635 KB631992 ENN79832.1 ERL87761.1 GBXI01016899 JAC97392.1 ATLV01020284 KE525297 KFB45016.1 GAKP01006771 GAKP01006770 JAC52182.1 GGMS01010625 MBY79828.1 AJVK01012925 CH933808 EDW10384.2 CH963849 EDW74386.1 AAAB01008900 EAA09395.4 APCN01002794 CAQQ02119555 CAQQ02119556 CAQQ02119557 CAQQ02119558 CAQQ02119559 CAQQ02119560 NNAY01002144 OXU21942.1 GBYB01012658 JAG82425.1 CM000071 EAL26068.1 AJWK01029481 AXCM01011569 CH940648 EDW61336.1 AB701637 BAL70289.1 CH916367 EDW01341.1 KK107353 QOIP01000012 EZA52081.1 RLU15762.1 LBMM01001031 KMQ97046.1 GAMC01002393 JAC04163.1 CH479181 EDW32526.1 BDGG01000001 GAU88503.1 CH477383 EAT42156.1 CP012524 ALC41286.1 KK107324 QOIP01000006 EZA52517.1 RLU21332.1 GEEE01000265 JAP62960.1 ADTU01010579 ADTU01010580 ADTU01010581 CP026247 AWP01225.1 KQ435794 KOX73598.1 EAAA01000909 APGK01008978 APGK01008979 APGK01008980 APGK01008981 KB737825 ENN82906.1 OUUW01000001 SPP73856.1 KQ435878 KOX69968.1 GFJQ02004357 JAW02613.1 KQ971372 EFA09563.1 GBYB01003736 JAG73503.1 ADMH02000552 ETN65900.1 UFQS01000997 UFQT01000997 SSX08337.1 SSX28360.1 GL451531 EFN79122.1 EFN79123.1 AAZX01003292 KQ976724 KYM76647.1 EFA09564.2 BX901962 CT573114 BC074038 BC142792 CH902619 EDV35587.1 AE013599 AAM68735.1 JTDY01005790 KOB66627.1 GL445031 EFN60280.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
UP000283053
+ More
UP000054359 UP000002358 UP000069272 UP000019118 UP000030742 UP000030765 UP000075900 UP000092462 UP000009192 UP000007798 UP000007062 UP000076407 UP000075840 UP000015102 UP000215335 UP000001819 UP000092461 UP000075883 UP000075920 UP000008792 UP000001070 UP000053097 UP000279307 UP000075885 UP000036403 UP000008744 UP000186922 UP000008820 UP000092553 UP000005226 UP000005205 UP000261540 UP000076408 UP000246464 UP000053105 UP000008144 UP000268350 UP000257200 UP000007266 UP000000673 UP000008237 UP000261500 UP000007875 UP000095300 UP000192223 UP000095301 UP000078540 UP000265140 UP000000437 UP000264820 UP000007801 UP000000803 UP000000311
UP000054359 UP000002358 UP000069272 UP000019118 UP000030742 UP000030765 UP000075900 UP000092462 UP000009192 UP000007798 UP000007062 UP000076407 UP000075840 UP000015102 UP000215335 UP000001819 UP000092461 UP000075883 UP000075920 UP000008792 UP000001070 UP000053097 UP000279307 UP000075885 UP000036403 UP000008744 UP000186922 UP000008820 UP000092553 UP000005226 UP000005205 UP000261540 UP000076408 UP000246464 UP000053105 UP000008144 UP000268350 UP000257200 UP000007266 UP000000673 UP000008237 UP000261500 UP000007875 UP000095300 UP000192223 UP000095301 UP000078540 UP000265140 UP000000437 UP000264820 UP000007801 UP000000803 UP000000311
PRIDE
Interpro
IPR018499
Tetraspanin/Peripherin
+ More
IPR000301 Tetraspanin
IPR008952 Tetraspanin_EC2_sf
IPR018503 Tetraspanin_CS
IPR024079 MetalloPept_cat_dom_sf
IPR008753 Peptidase_M13_N
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR042089 Peptidase_M13_dom_2
IPR036396 Cyt_P450_sf
IPR000718 Peptidase_M13
IPR018497 Peptidase_M13_C
IPR000301 Tetraspanin
IPR008952 Tetraspanin_EC2_sf
IPR018503 Tetraspanin_CS
IPR024079 MetalloPept_cat_dom_sf
IPR008753 Peptidase_M13_N
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR042089 Peptidase_M13_dom_2
IPR036396 Cyt_P450_sf
IPR000718 Peptidase_M13
IPR018497 Peptidase_M13_C
CDD
ProteinModelPortal
A0A2A4JFK4
A0A2H1W865
A0A212F783
A0A0L7KZ26
A0A194QRJ2
A0A194Q3L9
+ More
A0A212F781 A0A194Q457 A0A3S2P1W8 A0A3S2LJ58 A0A2A4JFM4 A0A212F793 A0A194Q9P1 A0A2H8TQA1 A0A2H1W9Q9 A0A087TFG6 A0A076KUR5 K7J9H7 A0A0B5H7U5 A0A182FCW0 A0A0K8WBF2 N6UM92 A0A0A1WG71 A0A084W472 A0A034WD08 A0A2S2QQ20 A0A182RV02 A0A1B0D9D5 B4KR28 B4MQJ7 Q7Q9H6 A0A182XET4 A0A182HNW1 T1GNT4 A0A232EUB7 A0A0C9QE23 Q28Y97 A0A1B0CUN9 A0A182MBF2 A0A182WME0 B4LQ51 H7CHJ1 B4J8Z2 A0A026W8X0 A0A182P8U7 A0A0J7L311 W8BXV6 B4GCW3 A0A1D1UQ65 Q176T5 A0A0M5J749 A0A026W8U6 A0A3B5KLI5 A0A0V0JAW3 A0A158NAZ0 A0A3B3RMI6 A0A182YC21 A0A2U9BBA8 A0A0N0BFU5 F6TTN6 N6UQ61 A0A3B0JKU2 A0A0N0BD54 A0A1Z5L5C1 H2UYH5 A0A3Q1FFI0 D6X0S1 A0A3B3RJZ9 A0A0C9RAA1 W5JSN2 A0A336MEI8 E2BYV0 A0A3B3ULN6 H2YU34 A0A1I8QDE5 K7ISS9 A0A1W4XT51 A0A1I8MII5 A0A151HY78 D6X0S0 A0A3P8YU62 Q6GMK6 A0A3B3RM52 A0A3Q2ZL31 B3MFB3 A1Z8K7 A0A0L7KTN2 E2B224
A0A212F781 A0A194Q457 A0A3S2P1W8 A0A3S2LJ58 A0A2A4JFM4 A0A212F793 A0A194Q9P1 A0A2H8TQA1 A0A2H1W9Q9 A0A087TFG6 A0A076KUR5 K7J9H7 A0A0B5H7U5 A0A182FCW0 A0A0K8WBF2 N6UM92 A0A0A1WG71 A0A084W472 A0A034WD08 A0A2S2QQ20 A0A182RV02 A0A1B0D9D5 B4KR28 B4MQJ7 Q7Q9H6 A0A182XET4 A0A182HNW1 T1GNT4 A0A232EUB7 A0A0C9QE23 Q28Y97 A0A1B0CUN9 A0A182MBF2 A0A182WME0 B4LQ51 H7CHJ1 B4J8Z2 A0A026W8X0 A0A182P8U7 A0A0J7L311 W8BXV6 B4GCW3 A0A1D1UQ65 Q176T5 A0A0M5J749 A0A026W8U6 A0A3B5KLI5 A0A0V0JAW3 A0A158NAZ0 A0A3B3RMI6 A0A182YC21 A0A2U9BBA8 A0A0N0BFU5 F6TTN6 N6UQ61 A0A3B0JKU2 A0A0N0BD54 A0A1Z5L5C1 H2UYH5 A0A3Q1FFI0 D6X0S1 A0A3B3RJZ9 A0A0C9RAA1 W5JSN2 A0A336MEI8 E2BYV0 A0A3B3ULN6 H2YU34 A0A1I8QDE5 K7ISS9 A0A1W4XT51 A0A1I8MII5 A0A151HY78 D6X0S0 A0A3P8YU62 Q6GMK6 A0A3B3RM52 A0A3Q2ZL31 B3MFB3 A1Z8K7 A0A0L7KTN2 E2B224
Ontologies
GO
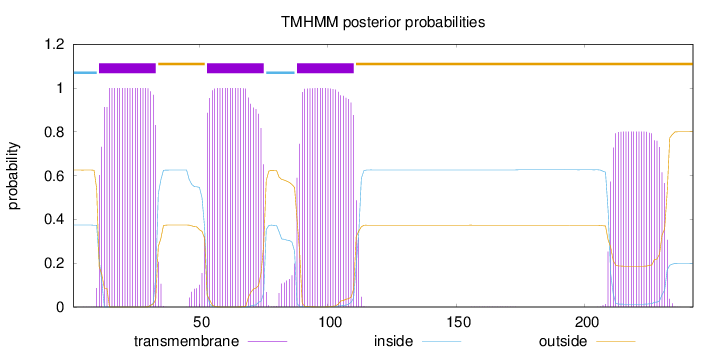
Topology
Subcellular location
Membrane
Length:
242
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
86.17253
Exp number, first 60 AAs:
31.03724
Total prob of N-in:
0.37452
POSSIBLE N-term signal
sequence
inside
1 - 10
TMhelix
11 - 33
outside
34 - 52
TMhelix
53 - 75
inside
76 - 87
TMhelix
88 - 110
outside
111 - 242
Population Genetic Test Statistics
Pi
131.854376
Theta
136.021641
Tajima's D
0.3985
CLR
0.883989
CSRT
0.490525473726314
Interpretation
Uncertain
