Gene
KWMTBOMO05940
Pre Gene Modal
BGIBMGA002910
Annotation
putative_sulfate/bicarbonate/oxalate_exchanger_sat-1_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.927
Sequence
CDS
ATGAAGACTTTGAAGAAACGGTTGCCGATCCTCGGTTGGTTACCCAACTATAAACCCGAATATTTCGTGCAAGACTTCATTGCCGGCATAACAGTGGGATTTACAGCCATACCGCAAGGCATAGCGTATGCTGTGGTGGCGGGGCAATCTCCGGAACATGGGCTCTATGCATCTCTGACTTCAGGAGTAATATACGTTATCTTCGGTAGCCGCTATAACGTGACTGTTGGGCCGACAGCTATCTTGGCAGCGATGACAGCAAAATACGTATCCGGCTATTCAGTGGATTTCGCAATTTTAACTGCGTTCGTTTCTGGTGTAGTGATTTTGTTGATGGGAGTGTTTCATCTCGGTTTTTTGGTTGAATTTATATCAATGCCGGTCATTAGTGGATTTACGACAGCCGCAGCCTTACAAATCGCTGCGTTACAACTAAAACCATTCTTCGGTCTCAGTGGATCATCCGGGAACAACTTTACCGAGTCCGTACAAAATCTAATACGTAATTTCAAAACTATGAAACTATACGAATTTCTTCTTGGATCAGCCACAATCATCATGTTGATGCTATTGAAAAAACTAGGTGATGGTTGCAAAAGAACTGACGGTTTATTTAAAAAAATACGATGGTTCATTTCGCTGTCTCGGAATGCGGTGGTTGTGATGGTGGGGATAACAGTAGCTTTTGTCATTAAAATTGTGTTTGATCAAGAACCATTAGAATTAATCGGTGCTATTGACGGTGGATTACCCAAATTCAGCTGGCCACCATTTTCTACAGTGGTCGGCAACGAAACATACAATTTTGGAGATATGATGGGCGTCTTAGGCCCCGGGTTTATAGTGCTACCGTTTGTTGCAATTATGGAAACGGTGGCCATCGCTAAAGCTTTTGCAGGAGGCGCCATAGTGGATGCCAATCAAGAAATGATCGCTCTTGGCTTGTGTAACATGATTGGATCTTTCGGACAAAGTATGCCAATCACAGGATCATTTACGAGGACGGCCATAAATCATATGTCTGGAGTTGTCACCCCACTGGGAGGTGTCACTAAAGTTCTTTTACTAGTTGTTGCGCTCACATCATTGACTTCAACGTTCTATTACATACCGAAAGCGTCTTTGGCCGGATTGATTATATCAGCAATGTTTTTCATGATCGATTATAAAACAATGCTGCAATTGTGGAGAAACAGCAAAAAAGAACTAGCTATCCTGCTCACGACCACATTTGTTTGTTTATCATTCGGTTTGGAATACGGCATCATAGCTGGAATTATCATCGAAGCCGCTTTGTTACTGCATAGAGTGTCAAGACCCAACCTGGGTGCTAACATAGTGAAATATCAGAAAGGCGATTTGCTTATTGTACCTTTCTCTGAAGATATATCGTATTGTGCCGCTGAACACGTTCGCAGAACAGTCATTAAAGAATCGCAAGAACTATCTGACACAGTAATCGTAATTGATGGTACGAACCTTAAAAACATGGATTTCACGGCTGCTTCCAATTTAGTATTGGTTGTAAAGGAGTTGGACACGAAATCCCTTACAGTAATTATGCTTAATTTTAATTCGACATTGAAGAAATTGTGTGTCGATATTGACAGAAGTACAGAAGACAAATTCGTTTACGCTAAAAACGCTATGGAAATATCAGAAATGAATCCCAGAGACGTACGTAGTAATGTTTAA
Protein
MKTLKKRLPILGWLPNYKPEYFVQDFIAGITVGFTAIPQGIAYAVVAGQSPEHGLYASLTSGVIYVIFGSRYNVTVGPTAILAAMTAKYVSGYSVDFAILTAFVSGVVILLMGVFHLGFLVEFISMPVISGFTTAAALQIAALQLKPFFGLSGSSGNNFTESVQNLIRNFKTMKLYEFLLGSATIIMLMLLKKLGDGCKRTDGLFKKIRWFISLSRNAVVVMVGITVAFVIKIVFDQEPLELIGAIDGGLPKFSWPPFSTVVGNETYNFGDMMGVLGPGFIVLPFVAIMETVAIAKAFAGGAIVDANQEMIALGLCNMIGSFGQSMPITGSFTRTAINHMSGVVTPLGGVTKVLLLVVALTSLTSTFYYIPKASLAGLIISAMFFMIDYKTMLQLWRNSKKELAILLTTTFVCLSFGLEYGIIAGIIIEAALLLHRVSRPNLGANIVKYQKGDLLIVPFSEDISYCAAEHVRRTVIKESQELSDTVIVIDGTNLKNMDFTAASNLVLVVKELDTKSLTVIMLNFNSTLKKLCVDIDRSTEDKFVYAKNAMEISEMNPRDVRSNV
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
H9J075
A0A0L7KNT3
A0A2H1W8A2
A0A2A4IUJ0
A0A194Q3K6
A0A194QRI6
+ More
A0A0L7LNJ8 A0A212EUS1 A0A224XKX7 A0A069DZB4 A0A1S4FV28 A0A182SIW7 A0A0V0G6Q9 Q16NA2 K7IZ33 A0A2H1V3R0 E2C1N4 A0A1L8DE51 A0A1Q3FHA0 A0A1L8DE43 A0A232EP62 A0A084W7X8 E2AIV7 F4WGT3 A0A2A4IZE9 A0A195BCI1 A0A232EQT5 K7IUX6 W5JR59 A0A2W1BLZ9 A0A195D872 A0A158NFQ0 A0A0P4VH77 A0A1Y1LG60 A0A1Y1LHP7 T1HYZ0 A0A146LXD8 A0A146M5W7 A0A084W7X7 A0A1B6GSE7 A0A1B6H7W1 A0A1B6GL11 A0A182G810 A0A182G9X9 A0A1B6HPI4 A0A1S4GLM3 E1ZW29 A0A2M4ARZ1 A0A151WPU2 A0A023EVF2 A0A1B6MPB0 A0A1B6M068 A0A182FP05 A0A151JSC4 A0A336KGN5 B0W0Z2 E2C4G6 W5JPJ6 A0A088A5S5 A0A182SBA7 A0A182PSS1 A0A336LHS8 A0A1I8PCV3 A0A1I8PD59 A0A182X5D1 A0A1I8PD52 A0A182VPV5 A0A139WCS7 A0A1Q3FH35 A0A182IDI4 A0A195FB80 A0A1B6CZ41 A0A1B6CXU3 A0A182NE09 A0A1B6E1K4 A0A2A3ENJ8 A0A182QWX7 A0A1B6EA75 A0A2J7RSI6 A0A1B6D085 A0A1B6DL52 A0A2A3E3G6 A0A1B6D0S6 A0A1B6D6E7 A0A1B6CM06 A0A0L0C6N7 A0A1W4X8R3 A0A087ZNT5 A0A1W4XIY6 A0A1B6JSW8 A0A2R7WJY0 A0A182K947 A0A0L7RA85 A0A182RE90 K7IYL9 B4J9J0 A0A182YER6 B4LKR8 F4X3H0
A0A0L7LNJ8 A0A212EUS1 A0A224XKX7 A0A069DZB4 A0A1S4FV28 A0A182SIW7 A0A0V0G6Q9 Q16NA2 K7IZ33 A0A2H1V3R0 E2C1N4 A0A1L8DE51 A0A1Q3FHA0 A0A1L8DE43 A0A232EP62 A0A084W7X8 E2AIV7 F4WGT3 A0A2A4IZE9 A0A195BCI1 A0A232EQT5 K7IUX6 W5JR59 A0A2W1BLZ9 A0A195D872 A0A158NFQ0 A0A0P4VH77 A0A1Y1LG60 A0A1Y1LHP7 T1HYZ0 A0A146LXD8 A0A146M5W7 A0A084W7X7 A0A1B6GSE7 A0A1B6H7W1 A0A1B6GL11 A0A182G810 A0A182G9X9 A0A1B6HPI4 A0A1S4GLM3 E1ZW29 A0A2M4ARZ1 A0A151WPU2 A0A023EVF2 A0A1B6MPB0 A0A1B6M068 A0A182FP05 A0A151JSC4 A0A336KGN5 B0W0Z2 E2C4G6 W5JPJ6 A0A088A5S5 A0A182SBA7 A0A182PSS1 A0A336LHS8 A0A1I8PCV3 A0A1I8PD59 A0A182X5D1 A0A1I8PD52 A0A182VPV5 A0A139WCS7 A0A1Q3FH35 A0A182IDI4 A0A195FB80 A0A1B6CZ41 A0A1B6CXU3 A0A182NE09 A0A1B6E1K4 A0A2A3ENJ8 A0A182QWX7 A0A1B6EA75 A0A2J7RSI6 A0A1B6D085 A0A1B6DL52 A0A2A3E3G6 A0A1B6D0S6 A0A1B6D6E7 A0A1B6CM06 A0A0L0C6N7 A0A1W4X8R3 A0A087ZNT5 A0A1W4XIY6 A0A1B6JSW8 A0A2R7WJY0 A0A182K947 A0A0L7RA85 A0A182RE90 K7IYL9 B4J9J0 A0A182YER6 B4LKR8 F4X3H0
Pubmed
EMBL
BABH01019037
JTDY01007741
KOB64973.1
ODYU01006956
SOQ49277.1
NWSH01007697
+ More
PCG62852.1 KQ459472 KPJ00123.1 KQ461175 KPJ07949.1 JTDY01000476 KOB77005.1 AGBW02012324 OWR45211.1 GFTR01007336 JAW09090.1 GBGD01000935 JAC87954.1 GECL01002483 JAP03641.1 CH477832 EAT35825.1 ODYU01000543 SOQ35478.1 GL451937 EFN78210.1 GFDF01009352 JAV04732.1 GFDL01008117 JAV26928.1 GFDF01009351 JAV04733.1 NNAY01003020 OXU20106.1 ATLV01021319 KE525316 KFB46322.1 GL439905 EFN66636.1 GL888147 EGI66552.1 NWSH01004247 PCG65307.1 KQ976514 KYM82271.1 NNAY01002700 OXU20749.1 AAZX01001196 AAZX01010281 ADMH02000728 ETN65234.1 KZ150001 PZC75311.1 KQ976750 KYN08644.1 ADTU01014484 ADTU01014485 ADTU01014486 ADTU01014487 ADTU01014488 ADTU01014489 GDKW01002824 JAI53771.1 GEZM01058159 JAV71861.1 GEZM01058160 JAV71860.1 ACPB03014185 GDHC01006325 JAQ12304.1 GDHC01003476 JAQ15153.1 KFB46321.1 GECZ01025058 GECZ01004421 JAS44711.1 JAS65348.1 GECU01037002 JAS70704.1 GECZ01021069 GECZ01006625 JAS48700.1 JAS63144.1 JXUM01047217 JXUM01047218 KQ561510 KXJ78346.1 JXUM01049949 JXUM01049950 GECU01031114 GECU01026481 JAS76592.1 JAS81225.1 AAAB01008952 GL434761 EFN74604.1 GGFK01010181 MBW43502.1 KQ982877 KYQ49705.1 GAPW01000485 JAC13113.1 GEBQ01002282 JAT37695.1 GEBQ01010697 JAT29280.1 KQ978538 KYN30256.1 UFQS01000397 UFQT01000397 SSX03619.1 SSX23984.1 DS231819 EDS42809.1 GL452471 EFN77150.1 ETN65233.1 UFQS01004118 UFQT01004118 SSX16183.1 SSX35509.1 KQ971362 KYB25749.1 GFDL01008179 JAV26866.1 APCN01005988 KQ981693 KYN37880.1 GEDC01018755 JAS18543.1 GEDC01019009 JAS18289.1 GEDC01005490 JAS31808.1 KZ288204 PBC33308.1 AXCN02002212 GEDC01002456 JAS34842.1 NEVH01000254 PNF43806.1 GEDC01018210 JAS19088.1 GEDC01010889 JAS26409.1 KZ288400 PBC26293.1 GEDC01018041 JAS19257.1 GEDC01016037 JAS21261.1 GEDC01022794 JAS14504.1 JRES01000835 KNC27930.1 GECU01005439 JAT02268.1 KK854946 PTY19868.1 KQ414619 KOC67754.1 CH916367 EDW02497.1 CH940648 EDW61791.2 GL888613 EGI58998.1
PCG62852.1 KQ459472 KPJ00123.1 KQ461175 KPJ07949.1 JTDY01000476 KOB77005.1 AGBW02012324 OWR45211.1 GFTR01007336 JAW09090.1 GBGD01000935 JAC87954.1 GECL01002483 JAP03641.1 CH477832 EAT35825.1 ODYU01000543 SOQ35478.1 GL451937 EFN78210.1 GFDF01009352 JAV04732.1 GFDL01008117 JAV26928.1 GFDF01009351 JAV04733.1 NNAY01003020 OXU20106.1 ATLV01021319 KE525316 KFB46322.1 GL439905 EFN66636.1 GL888147 EGI66552.1 NWSH01004247 PCG65307.1 KQ976514 KYM82271.1 NNAY01002700 OXU20749.1 AAZX01001196 AAZX01010281 ADMH02000728 ETN65234.1 KZ150001 PZC75311.1 KQ976750 KYN08644.1 ADTU01014484 ADTU01014485 ADTU01014486 ADTU01014487 ADTU01014488 ADTU01014489 GDKW01002824 JAI53771.1 GEZM01058159 JAV71861.1 GEZM01058160 JAV71860.1 ACPB03014185 GDHC01006325 JAQ12304.1 GDHC01003476 JAQ15153.1 KFB46321.1 GECZ01025058 GECZ01004421 JAS44711.1 JAS65348.1 GECU01037002 JAS70704.1 GECZ01021069 GECZ01006625 JAS48700.1 JAS63144.1 JXUM01047217 JXUM01047218 KQ561510 KXJ78346.1 JXUM01049949 JXUM01049950 GECU01031114 GECU01026481 JAS76592.1 JAS81225.1 AAAB01008952 GL434761 EFN74604.1 GGFK01010181 MBW43502.1 KQ982877 KYQ49705.1 GAPW01000485 JAC13113.1 GEBQ01002282 JAT37695.1 GEBQ01010697 JAT29280.1 KQ978538 KYN30256.1 UFQS01000397 UFQT01000397 SSX03619.1 SSX23984.1 DS231819 EDS42809.1 GL452471 EFN77150.1 ETN65233.1 UFQS01004118 UFQT01004118 SSX16183.1 SSX35509.1 KQ971362 KYB25749.1 GFDL01008179 JAV26866.1 APCN01005988 KQ981693 KYN37880.1 GEDC01018755 JAS18543.1 GEDC01019009 JAS18289.1 GEDC01005490 JAS31808.1 KZ288204 PBC33308.1 AXCN02002212 GEDC01002456 JAS34842.1 NEVH01000254 PNF43806.1 GEDC01018210 JAS19088.1 GEDC01010889 JAS26409.1 KZ288400 PBC26293.1 GEDC01018041 JAS19257.1 GEDC01016037 JAS21261.1 GEDC01022794 JAS14504.1 JRES01000835 KNC27930.1 GECU01005439 JAT02268.1 KK854946 PTY19868.1 KQ414619 KOC67754.1 CH916367 EDW02497.1 CH940648 EDW61791.2 GL888613 EGI58998.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000075901 UP000008820 UP000002358 UP000008237 UP000215335 UP000030765 UP000000311 UP000007755 UP000078540 UP000000673 UP000078542 UP000005205 UP000015103 UP000069940 UP000249989 UP000075809 UP000069272 UP000078492 UP000002320 UP000005203 UP000075885 UP000095300 UP000076407 UP000075920 UP000007266 UP000075840 UP000078541 UP000075884 UP000242457 UP000075886 UP000235965 UP000037069 UP000192223 UP000075881 UP000053825 UP000075900 UP000001070 UP000076408 UP000008792
UP000075901 UP000008820 UP000002358 UP000008237 UP000215335 UP000030765 UP000000311 UP000007755 UP000078540 UP000000673 UP000078542 UP000005205 UP000015103 UP000069940 UP000249989 UP000075809 UP000069272 UP000078492 UP000002320 UP000005203 UP000075885 UP000095300 UP000076407 UP000075920 UP000007266 UP000075840 UP000078541 UP000075884 UP000242457 UP000075886 UP000235965 UP000037069 UP000192223 UP000075881 UP000053825 UP000075900 UP000001070 UP000076408 UP000008792
PRIDE
Interpro
SUPFAM
SSF52091
SSF52091
Gene 3D
ProteinModelPortal
H9J075
A0A0L7KNT3
A0A2H1W8A2
A0A2A4IUJ0
A0A194Q3K6
A0A194QRI6
+ More
A0A0L7LNJ8 A0A212EUS1 A0A224XKX7 A0A069DZB4 A0A1S4FV28 A0A182SIW7 A0A0V0G6Q9 Q16NA2 K7IZ33 A0A2H1V3R0 E2C1N4 A0A1L8DE51 A0A1Q3FHA0 A0A1L8DE43 A0A232EP62 A0A084W7X8 E2AIV7 F4WGT3 A0A2A4IZE9 A0A195BCI1 A0A232EQT5 K7IUX6 W5JR59 A0A2W1BLZ9 A0A195D872 A0A158NFQ0 A0A0P4VH77 A0A1Y1LG60 A0A1Y1LHP7 T1HYZ0 A0A146LXD8 A0A146M5W7 A0A084W7X7 A0A1B6GSE7 A0A1B6H7W1 A0A1B6GL11 A0A182G810 A0A182G9X9 A0A1B6HPI4 A0A1S4GLM3 E1ZW29 A0A2M4ARZ1 A0A151WPU2 A0A023EVF2 A0A1B6MPB0 A0A1B6M068 A0A182FP05 A0A151JSC4 A0A336KGN5 B0W0Z2 E2C4G6 W5JPJ6 A0A088A5S5 A0A182SBA7 A0A182PSS1 A0A336LHS8 A0A1I8PCV3 A0A1I8PD59 A0A182X5D1 A0A1I8PD52 A0A182VPV5 A0A139WCS7 A0A1Q3FH35 A0A182IDI4 A0A195FB80 A0A1B6CZ41 A0A1B6CXU3 A0A182NE09 A0A1B6E1K4 A0A2A3ENJ8 A0A182QWX7 A0A1B6EA75 A0A2J7RSI6 A0A1B6D085 A0A1B6DL52 A0A2A3E3G6 A0A1B6D0S6 A0A1B6D6E7 A0A1B6CM06 A0A0L0C6N7 A0A1W4X8R3 A0A087ZNT5 A0A1W4XIY6 A0A1B6JSW8 A0A2R7WJY0 A0A182K947 A0A0L7RA85 A0A182RE90 K7IYL9 B4J9J0 A0A182YER6 B4LKR8 F4X3H0
A0A0L7LNJ8 A0A212EUS1 A0A224XKX7 A0A069DZB4 A0A1S4FV28 A0A182SIW7 A0A0V0G6Q9 Q16NA2 K7IZ33 A0A2H1V3R0 E2C1N4 A0A1L8DE51 A0A1Q3FHA0 A0A1L8DE43 A0A232EP62 A0A084W7X8 E2AIV7 F4WGT3 A0A2A4IZE9 A0A195BCI1 A0A232EQT5 K7IUX6 W5JR59 A0A2W1BLZ9 A0A195D872 A0A158NFQ0 A0A0P4VH77 A0A1Y1LG60 A0A1Y1LHP7 T1HYZ0 A0A146LXD8 A0A146M5W7 A0A084W7X7 A0A1B6GSE7 A0A1B6H7W1 A0A1B6GL11 A0A182G810 A0A182G9X9 A0A1B6HPI4 A0A1S4GLM3 E1ZW29 A0A2M4ARZ1 A0A151WPU2 A0A023EVF2 A0A1B6MPB0 A0A1B6M068 A0A182FP05 A0A151JSC4 A0A336KGN5 B0W0Z2 E2C4G6 W5JPJ6 A0A088A5S5 A0A182SBA7 A0A182PSS1 A0A336LHS8 A0A1I8PCV3 A0A1I8PD59 A0A182X5D1 A0A1I8PD52 A0A182VPV5 A0A139WCS7 A0A1Q3FH35 A0A182IDI4 A0A195FB80 A0A1B6CZ41 A0A1B6CXU3 A0A182NE09 A0A1B6E1K4 A0A2A3ENJ8 A0A182QWX7 A0A1B6EA75 A0A2J7RSI6 A0A1B6D085 A0A1B6DL52 A0A2A3E3G6 A0A1B6D0S6 A0A1B6D6E7 A0A1B6CM06 A0A0L0C6N7 A0A1W4X8R3 A0A087ZNT5 A0A1W4XIY6 A0A1B6JSW8 A0A2R7WJY0 A0A182K947 A0A0L7RA85 A0A182RE90 K7IYL9 B4J9J0 A0A182YER6 B4LKR8 F4X3H0
PDB
5DA0
E-value=1.72486e-09,
Score=151
Ontologies
GO
PANTHER
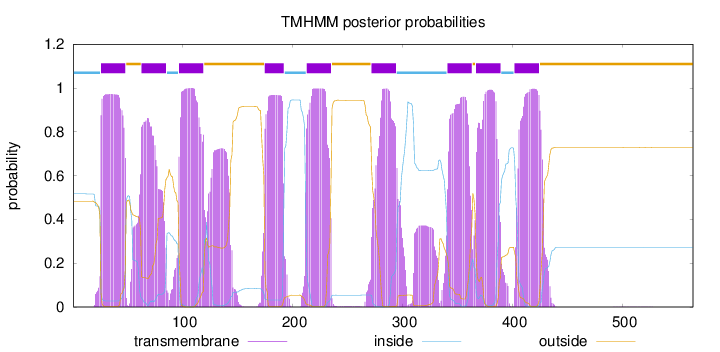
Topology
Subcellular location
Membrane
Length:
564
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
216.38764
Exp number, first 60 AAs:
24.58318
Total prob of N-in:
0.51667
POSSIBLE N-term signal
sequence
inside
1 - 25
TMhelix
26 - 48
outside
49 - 62
TMhelix
63 - 85
inside
86 - 96
TMhelix
97 - 119
outside
120 - 174
TMhelix
175 - 192
inside
193 - 212
TMhelix
213 - 235
outside
236 - 271
TMhelix
272 - 294
inside
295 - 340
TMhelix
341 - 363
outside
364 - 366
TMhelix
367 - 389
inside
390 - 401
TMhelix
402 - 424
outside
425 - 564
Population Genetic Test Statistics
Pi
255.932928
Theta
219.031922
Tajima's D
0.87712
CLR
0
CSRT
0.625768711564422
Interpretation
Uncertain
