Pre Gene Modal
BGIBMGA002793
Annotation
endoplasmic_reticulum_oxidoreduction_1-like_precursor_[Bombyx_mori]
Full name
Ero1-like protein
Alternative Name
Endoplasmic reticulum oxidoreductin-1-like protein
Location in the cell
Extracellular Reliability : 1.702 Nuclear Reliability : 1.718
Sequence
CDS
ATGGCGCGGACATCCACAGAGATGCCACAGAAAAGTTATTGTGTTGTGTTTATTTTATTTGCTCTAGCGATAGTACAGGCGGTTGGTTACGACACGGAGCTAACGAAGCCTTGCGACAGTACAGCCTGCTTCGATGAGCTCCACGGGTCCTTGGGGGACTGCTCCTGTAAAGTCGACACAATTGACTTCTTCAATAATGTGAAGATCTTTCCCCGCATCCAGAGTATAGTCACGAAAGACTACTTCCGTTTTTATAAGGTTAATTTAAAAAAGGAATGCCCGTTCTGGGCTGATGACAGTAGGTGTGCAATGAAATACTGCCATATAAAGACTTGCACGAAGGAAAGTGTACCTGGCTATGAAACTGGATATAATAATGAGTATGTAGATGAAACGCCTGCCACCAAATACTCTAAAGAAGCACAGTCCGACTGTAAGTCTGATTTGGATCACGATCCTCAGTTGAGCTATTTGAATATGACCATTAGTGCTGCCAACCAGTATGAAATAGCAAAATGGAAAGTTCACGATGATGCACATGATAATTTTTGTGAAAGTGATGACCGTGATGCTGATGCAGATTATGTTGATTTAACATTAAACCCAGAGAGGTATACAGGATATAAAGGGCCATCTGCACACAGAATTTGGAGGAGTATTTATCAAGAAAACTGCTTTCGTCCAAAATTAAATCCATACGAATCATTCCCTTATGTGTTAAGTTCAGATCTGAGTAACCTGTGTTTAGAAAAACGAGTGTTTTATAGAGCTATATCTGGACTCCATACAAGTATTAATATACATTTATGTTCAAAATTTCTTTTGTCAGAAAAAAGCTTAGGTTTTGTTGCTCCTCCGGAAGGAGAGTGGGGTCCAAACTTAGAAGAGTTTCAAAGGAGATTTGATCCCTCTCAAACTATGGGTGAAAGTACAAATTGGTTAAAAAATCTATATTTTGTGTACTTATTAGAAATGAGGGCATTAGCAAAAGCTGGGCCATATTTAGAACAAGAAGAATATTTTACGGGAAACCCAACAGAAGATGAAGAGACCCGACAGGCGATCAGCAACATGTTGGGTGTTATATACTCATTTCCTGAGCATTTCAATGAGTCTTCCATGTTCAAAGGAGGCAGCCAAGTGGAGACACTGAAAAATGAGTTTCGTGATCATTTCCGTAATATTTCCAGGATCATGGACTGTGTTGGCTGTGATAAATGCAAGTTGTGGGGTAAACTCCAAACTCAGGGTCTCGGGACTGCTTTGAAAATATTGTTCTCGGATAAATGGAATAGCCCCGAAGCAGATCCCTCAAGTGGGAAACTACCATTAAGACATAAGTCACATAAAAAGCTACAAAGAACAGAGATTGTGGCCCTGTTCAATGCATTTGCTAGATTATCAAACAGTATTCGAGAACTGGAAAATTTCAGAATCATGCTTAGGTCGAATCGAGATGAAAGTTTATTTGTCACGTGCCAGCTTATTGAAGTGTAA
Protein
MARTSTEMPQKSYCVVFILFALAIVQAVGYDTELTKPCDSTACFDELHGSLGDCSCKVDTIDFFNNVKIFPRIQSIVTKDYFRFYKVNLKKECPFWADDSRCAMKYCHIKTCTKESVPGYETGYNNEYVDETPATKYSKEAQSDCKSDLDHDPQLSYLNMTISAANQYEIAKWKVHDDAHDNFCESDDRDADADYVDLTLNPERYTGYKGPSAHRIWRSIYQENCFRPKLNPYESFPYVLSSDLSNLCLEKRVFYRAISGLHTSINIHLCSKFLLSEKSLGFVAPPEGEWGPNLEEFQRRFDPSQTMGESTNWLKNLYFVYLLEMRALAKAGPYLEQEEYFTGNPTEDEETRQAISNMLGVIYSFPEHFNESSMFKGGSQVETLKNEFRDHFRNISRIMDCVGCDKCKLWGKLQTQGLGTALKILFSDKWNSPEADPSSGKLPLRHKSHKKLQRTEIVALFNAFARLSNSIRELENFRIMLRSNRDESLFVTCQLIEV
Summary
Description
Oxidoreductase involved in disulfide bond formation in the endoplasmic reticulum. Efficiently reoxidizes pdi-1, the enzyme catalyzing protein disulfide formation, in order to allow pdi-1 to sustain additional rounds of disulfide formation. Following pdi reoxidation, passes its electrons to molecular oxygen via FAD, leading to the production of reactive oxygen species (ROS) in the cell (By similarity).
Cofactor
FAD
Subunit
May function both as a monomer and a homodimer.
Similarity
Belongs to the EROs family.
Keywords
Complete proteome
Disulfide bond
Electron transport
Endoplasmic reticulum
FAD
Flavoprotein
Glycoprotein
Membrane
Oxidoreductase
Redox-active center
Reference proteome
Signal
Transport
Feature
chain Ero1-like protein
Uniprot
B8Y9B6
B8Y9B7
H9IZV8
A0A3S2P8J9
A0A0L7L5B5
A0A2H1X222
+ More
A0A212EWW5 A0A194QR25 A0A194Q447 A0A2A4IUV4 A0A2J7QT53 A0A2J7QT51 A0A232F337 A0A0N0U3V4 A0A088AUZ0 K7ISD7 A0A2A3E833 A0A1B6C188 E2BCF6 A0A158N925 A0A195FTI4 A0A026VVV6 A0A151J7P6 A0A151WNQ0 F4WHH6 A0A151I6X8 E0VC42 A0A1B6FD23 A0A1B6IJN4 T1HPH1 A0A0P4VV13 E2A327 A0A224XP88 A0A0C9PTU8 D6W7V7 U4U9G2 N6TSI6 A0A0A9VX61 A0A0K8TPM8 A0A0A9VYW6 A0A0A9W2U9 A0A0K8SIA3 A0A023EU82 Q17J67 A0A1S4F0Q7 A0A336MIC1 A0A1Q3FFH4 A0A336MHH0 B0WB59 U5ETA6 A0A1Q3FFD8 U5EX41 A0A182G527 A0A0L7QL43 A0A0L0CDR1 A0A1L8DUA8 A0A182G4I8 A0A0K8W7W4 A0A1B0CX39 A0A034VHT1 B4PHQ6 A0A182LYV0 A0A0A1WIB5 A0A3B0JBM9 A0A182TPY3 A0A293MVS8 Q7QCV3 D6WXL9 Q9V3A6 A0A182XHQ5 A0A182USK6 A0A182LGI4 A0A182HTN3 B4LB92 B4L9D2 W8BLB0 A0A2H8TWB3 B3M4W0 A0A182W002 A0A182QL73 A0A195BVQ4 B4J1F1 A0A0J9RNR9 A0A182P271 A0A1Y1MUH4 B4H7V7 B5DRX3 B4HU11 A0A182RNX4 B3NG46 A0A182IWA6 A0A1A9WLK3 A0A1I8QCH5 A0A182N775 A0A084VZZ7 A0A182KCS8 A0A0G2YI10 A0A1A9YIH1 A0A182Y448 A0A2M4BLU4
A0A212EWW5 A0A194QR25 A0A194Q447 A0A2A4IUV4 A0A2J7QT53 A0A2J7QT51 A0A232F337 A0A0N0U3V4 A0A088AUZ0 K7ISD7 A0A2A3E833 A0A1B6C188 E2BCF6 A0A158N925 A0A195FTI4 A0A026VVV6 A0A151J7P6 A0A151WNQ0 F4WHH6 A0A151I6X8 E0VC42 A0A1B6FD23 A0A1B6IJN4 T1HPH1 A0A0P4VV13 E2A327 A0A224XP88 A0A0C9PTU8 D6W7V7 U4U9G2 N6TSI6 A0A0A9VX61 A0A0K8TPM8 A0A0A9VYW6 A0A0A9W2U9 A0A0K8SIA3 A0A023EU82 Q17J67 A0A1S4F0Q7 A0A336MIC1 A0A1Q3FFH4 A0A336MHH0 B0WB59 U5ETA6 A0A1Q3FFD8 U5EX41 A0A182G527 A0A0L7QL43 A0A0L0CDR1 A0A1L8DUA8 A0A182G4I8 A0A0K8W7W4 A0A1B0CX39 A0A034VHT1 B4PHQ6 A0A182LYV0 A0A0A1WIB5 A0A3B0JBM9 A0A182TPY3 A0A293MVS8 Q7QCV3 D6WXL9 Q9V3A6 A0A182XHQ5 A0A182USK6 A0A182LGI4 A0A182HTN3 B4LB92 B4L9D2 W8BLB0 A0A2H8TWB3 B3M4W0 A0A182W002 A0A182QL73 A0A195BVQ4 B4J1F1 A0A0J9RNR9 A0A182P271 A0A1Y1MUH4 B4H7V7 B5DRX3 B4HU11 A0A182RNX4 B3NG46 A0A182IWA6 A0A1A9WLK3 A0A1I8QCH5 A0A182N775 A0A084VZZ7 A0A182KCS8 A0A0G2YI10 A0A1A9YIH1 A0A182Y448 A0A2M4BLU4
EC Number
1.8.4.-
Pubmed
26556347
19121390
26227816
22118469
26354079
28648823
+ More
20075255 20798317 21347285 24508170 30249741 21719571 20566863 27129103 18362917 19820115 23537049 25401762 26823975 26369729 24945155 17510324 26483478 26108605 25348373 17994087 17550304 25830018 12364791 10671517 10731132 12537572 20966253 24495485 22936249 28004739 15632085 24438588 25244985
20075255 20798317 21347285 24508170 30249741 21719571 20566863 27129103 18362917 19820115 23537049 25401762 26823975 26369729 24945155 17510324 26483478 26108605 25348373 17994087 17550304 25830018 12364791 10671517 10731132 12537572 20966253 24495485 22936249 28004739 15632085 24438588 25244985
EMBL
FJ502246
ACL54850.1
FJ502247
ACL54851.1
BABH01019060
RSAL01000002
+ More
RVE55000.1 RVE55053.1 JTDY01002812 KOB70673.1 ODYU01012861 SOQ59371.1 AGBW02011867 OWR45998.1 KQ461175 KPJ07947.1 KQ459472 KPJ00119.1 NWSH01006341 PCG63501.1 NEVH01011202 PNF31772.1 PNF31771.1 NNAY01001180 OXU24837.1 KQ435883 KOX69758.1 KZ288356 PBC27211.1 GEDC01030628 GEDC01030031 GEDC01025685 GEDC01016183 GEDC01005881 GEDC01002076 JAS06670.1 JAS07267.1 JAS11613.1 JAS21115.1 JAS31417.1 JAS35222.1 GL447257 EFN86692.1 ADTU01009051 KQ981268 KYN43930.1 KK107785 QOIP01000004 EZA47800.1 RLU24120.1 KQ979640 KYN20038.1 KQ982907 KYQ49473.1 GL888161 EGI66330.1 KQ978451 KYM93926.1 DS235047 EEB10948.1 GECZ01021674 JAS48095.1 GECU01020581 JAS87125.1 ACPB03017117 GDKW01001943 JAI54652.1 GL436266 EFN72162.1 GFTR01006585 JAW09841.1 GBYB01004758 JAG74525.1 KQ971307 EFA11035.1 KB632243 ERL90554.1 APGK01057129 KB741277 ENN71296.1 GBHO01042807 GDHC01015422 GDHC01015232 JAG00797.1 JAQ03207.1 JAQ03397.1 GDAI01001246 JAI16357.1 GBHO01042805 JAG00799.1 GBHO01042806 GBRD01012784 JAG00798.1 JAG53042.1 GBRD01012788 JAG53038.1 GAPW01001132 JAC12466.1 CH477234 EAT46740.1 UFQT01001318 SSX30016.1 GFDL01008752 JAV26293.1 UFQS01001286 UFQT01001286 SSX10122.1 SSX29844.1 DS231877 EDS42124.1 GANO01002008 JAB57863.1 GFDL01008782 JAV26263.1 GANO01002690 JAB57181.1 JXUM01042865 KQ561341 KXJ78916.1 KQ414934 KOC59304.1 JRES01000645 KNC29629.1 GFDF01004056 JAV10028.1 JXUM01042515 JXUM01042516 KQ561327 KXJ78973.1 GDHF01005051 JAI47263.1 AJWK01033182 GAKP01017285 JAC41667.1 CM000159 EDW93365.1 AXCM01001634 GBXI01016154 JAC98137.1 OUUW01000002 SPP77442.1 GFWV01019491 MAA44219.1 AAAB01008859 EAA07773.5 KQ971362 EFA08867.1 AF125280 AE014296 BT021422 APCN01000570 CH940647 EDW68656.1 CH933816 EDW17307.1 GAMC01008752 JAB97803.1 GFXV01006770 MBW18575.1 CH902618 EDV40534.1 AXCN02000328 KQ976405 KYM91876.1 CH916366 EDV95842.1 CM002912 KMY97518.1 GEZM01020390 JAV89281.1 CH479219 EDW34747.1 CH379070 EDY74250.1 CH480817 EDW50432.1 CH954178 EDV50875.1 ATLV01019059 KE525259 KFB43541.1 KP322129 AKI88008.1 GGFJ01004861 MBW54002.1
RVE55000.1 RVE55053.1 JTDY01002812 KOB70673.1 ODYU01012861 SOQ59371.1 AGBW02011867 OWR45998.1 KQ461175 KPJ07947.1 KQ459472 KPJ00119.1 NWSH01006341 PCG63501.1 NEVH01011202 PNF31772.1 PNF31771.1 NNAY01001180 OXU24837.1 KQ435883 KOX69758.1 KZ288356 PBC27211.1 GEDC01030628 GEDC01030031 GEDC01025685 GEDC01016183 GEDC01005881 GEDC01002076 JAS06670.1 JAS07267.1 JAS11613.1 JAS21115.1 JAS31417.1 JAS35222.1 GL447257 EFN86692.1 ADTU01009051 KQ981268 KYN43930.1 KK107785 QOIP01000004 EZA47800.1 RLU24120.1 KQ979640 KYN20038.1 KQ982907 KYQ49473.1 GL888161 EGI66330.1 KQ978451 KYM93926.1 DS235047 EEB10948.1 GECZ01021674 JAS48095.1 GECU01020581 JAS87125.1 ACPB03017117 GDKW01001943 JAI54652.1 GL436266 EFN72162.1 GFTR01006585 JAW09841.1 GBYB01004758 JAG74525.1 KQ971307 EFA11035.1 KB632243 ERL90554.1 APGK01057129 KB741277 ENN71296.1 GBHO01042807 GDHC01015422 GDHC01015232 JAG00797.1 JAQ03207.1 JAQ03397.1 GDAI01001246 JAI16357.1 GBHO01042805 JAG00799.1 GBHO01042806 GBRD01012784 JAG00798.1 JAG53042.1 GBRD01012788 JAG53038.1 GAPW01001132 JAC12466.1 CH477234 EAT46740.1 UFQT01001318 SSX30016.1 GFDL01008752 JAV26293.1 UFQS01001286 UFQT01001286 SSX10122.1 SSX29844.1 DS231877 EDS42124.1 GANO01002008 JAB57863.1 GFDL01008782 JAV26263.1 GANO01002690 JAB57181.1 JXUM01042865 KQ561341 KXJ78916.1 KQ414934 KOC59304.1 JRES01000645 KNC29629.1 GFDF01004056 JAV10028.1 JXUM01042515 JXUM01042516 KQ561327 KXJ78973.1 GDHF01005051 JAI47263.1 AJWK01033182 GAKP01017285 JAC41667.1 CM000159 EDW93365.1 AXCM01001634 GBXI01016154 JAC98137.1 OUUW01000002 SPP77442.1 GFWV01019491 MAA44219.1 AAAB01008859 EAA07773.5 KQ971362 EFA08867.1 AF125280 AE014296 BT021422 APCN01000570 CH940647 EDW68656.1 CH933816 EDW17307.1 GAMC01008752 JAB97803.1 GFXV01006770 MBW18575.1 CH902618 EDV40534.1 AXCN02000328 KQ976405 KYM91876.1 CH916366 EDV95842.1 CM002912 KMY97518.1 GEZM01020390 JAV89281.1 CH479219 EDW34747.1 CH379070 EDY74250.1 CH480817 EDW50432.1 CH954178 EDV50875.1 ATLV01019059 KE525259 KFB43541.1 KP322129 AKI88008.1 GGFJ01004861 MBW54002.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000218220 UP000235965 UP000215335 UP000053105 UP000005203 UP000002358 UP000242457 UP000008237 UP000005205 UP000078541 UP000053097 UP000279307 UP000078492 UP000075809 UP000007755 UP000078542 UP000009046 UP000015103 UP000000311 UP000007266 UP000030742 UP000019118 UP000008820 UP000002320 UP000069940 UP000249989 UP000053825 UP000037069 UP000092461 UP000002282 UP000075883 UP000268350 UP000075902 UP000007062 UP000000803 UP000076407 UP000075903 UP000075882 UP000075840 UP000008792 UP000009192 UP000007801 UP000075920 UP000075886 UP000078540 UP000001070 UP000075885 UP000008744 UP000001819 UP000001292 UP000075900 UP000008711 UP000075880 UP000091820 UP000095300 UP000075884 UP000030765 UP000075881 UP000092443 UP000076408
UP000218220 UP000235965 UP000215335 UP000053105 UP000005203 UP000002358 UP000242457 UP000008237 UP000005205 UP000078541 UP000053097 UP000279307 UP000078492 UP000075809 UP000007755 UP000078542 UP000009046 UP000015103 UP000000311 UP000007266 UP000030742 UP000019118 UP000008820 UP000002320 UP000069940 UP000249989 UP000053825 UP000037069 UP000092461 UP000002282 UP000075883 UP000268350 UP000075902 UP000007062 UP000000803 UP000076407 UP000075903 UP000075882 UP000075840 UP000008792 UP000009192 UP000007801 UP000075920 UP000075886 UP000078540 UP000001070 UP000075885 UP000008744 UP000001819 UP000001292 UP000075900 UP000008711 UP000075880 UP000091820 UP000095300 UP000075884 UP000030765 UP000075881 UP000092443 UP000076408
Pfam
PF04137 ERO1
SUPFAM
SSF110019
SSF110019
ProteinModelPortal
B8Y9B6
B8Y9B7
H9IZV8
A0A3S2P8J9
A0A0L7L5B5
A0A2H1X222
+ More
A0A212EWW5 A0A194QR25 A0A194Q447 A0A2A4IUV4 A0A2J7QT53 A0A2J7QT51 A0A232F337 A0A0N0U3V4 A0A088AUZ0 K7ISD7 A0A2A3E833 A0A1B6C188 E2BCF6 A0A158N925 A0A195FTI4 A0A026VVV6 A0A151J7P6 A0A151WNQ0 F4WHH6 A0A151I6X8 E0VC42 A0A1B6FD23 A0A1B6IJN4 T1HPH1 A0A0P4VV13 E2A327 A0A224XP88 A0A0C9PTU8 D6W7V7 U4U9G2 N6TSI6 A0A0A9VX61 A0A0K8TPM8 A0A0A9VYW6 A0A0A9W2U9 A0A0K8SIA3 A0A023EU82 Q17J67 A0A1S4F0Q7 A0A336MIC1 A0A1Q3FFH4 A0A336MHH0 B0WB59 U5ETA6 A0A1Q3FFD8 U5EX41 A0A182G527 A0A0L7QL43 A0A0L0CDR1 A0A1L8DUA8 A0A182G4I8 A0A0K8W7W4 A0A1B0CX39 A0A034VHT1 B4PHQ6 A0A182LYV0 A0A0A1WIB5 A0A3B0JBM9 A0A182TPY3 A0A293MVS8 Q7QCV3 D6WXL9 Q9V3A6 A0A182XHQ5 A0A182USK6 A0A182LGI4 A0A182HTN3 B4LB92 B4L9D2 W8BLB0 A0A2H8TWB3 B3M4W0 A0A182W002 A0A182QL73 A0A195BVQ4 B4J1F1 A0A0J9RNR9 A0A182P271 A0A1Y1MUH4 B4H7V7 B5DRX3 B4HU11 A0A182RNX4 B3NG46 A0A182IWA6 A0A1A9WLK3 A0A1I8QCH5 A0A182N775 A0A084VZZ7 A0A182KCS8 A0A0G2YI10 A0A1A9YIH1 A0A182Y448 A0A2M4BLU4
A0A212EWW5 A0A194QR25 A0A194Q447 A0A2A4IUV4 A0A2J7QT53 A0A2J7QT51 A0A232F337 A0A0N0U3V4 A0A088AUZ0 K7ISD7 A0A2A3E833 A0A1B6C188 E2BCF6 A0A158N925 A0A195FTI4 A0A026VVV6 A0A151J7P6 A0A151WNQ0 F4WHH6 A0A151I6X8 E0VC42 A0A1B6FD23 A0A1B6IJN4 T1HPH1 A0A0P4VV13 E2A327 A0A224XP88 A0A0C9PTU8 D6W7V7 U4U9G2 N6TSI6 A0A0A9VX61 A0A0K8TPM8 A0A0A9VYW6 A0A0A9W2U9 A0A0K8SIA3 A0A023EU82 Q17J67 A0A1S4F0Q7 A0A336MIC1 A0A1Q3FFH4 A0A336MHH0 B0WB59 U5ETA6 A0A1Q3FFD8 U5EX41 A0A182G527 A0A0L7QL43 A0A0L0CDR1 A0A1L8DUA8 A0A182G4I8 A0A0K8W7W4 A0A1B0CX39 A0A034VHT1 B4PHQ6 A0A182LYV0 A0A0A1WIB5 A0A3B0JBM9 A0A182TPY3 A0A293MVS8 Q7QCV3 D6WXL9 Q9V3A6 A0A182XHQ5 A0A182USK6 A0A182LGI4 A0A182HTN3 B4LB92 B4L9D2 W8BLB0 A0A2H8TWB3 B3M4W0 A0A182W002 A0A182QL73 A0A195BVQ4 B4J1F1 A0A0J9RNR9 A0A182P271 A0A1Y1MUH4 B4H7V7 B5DRX3 B4HU11 A0A182RNX4 B3NG46 A0A182IWA6 A0A1A9WLK3 A0A1I8QCH5 A0A182N775 A0A084VZZ7 A0A182KCS8 A0A0G2YI10 A0A1A9YIH1 A0A182Y448 A0A2M4BLU4
PDB
3AHQ
E-value=1.14319e-110,
Score=1023
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
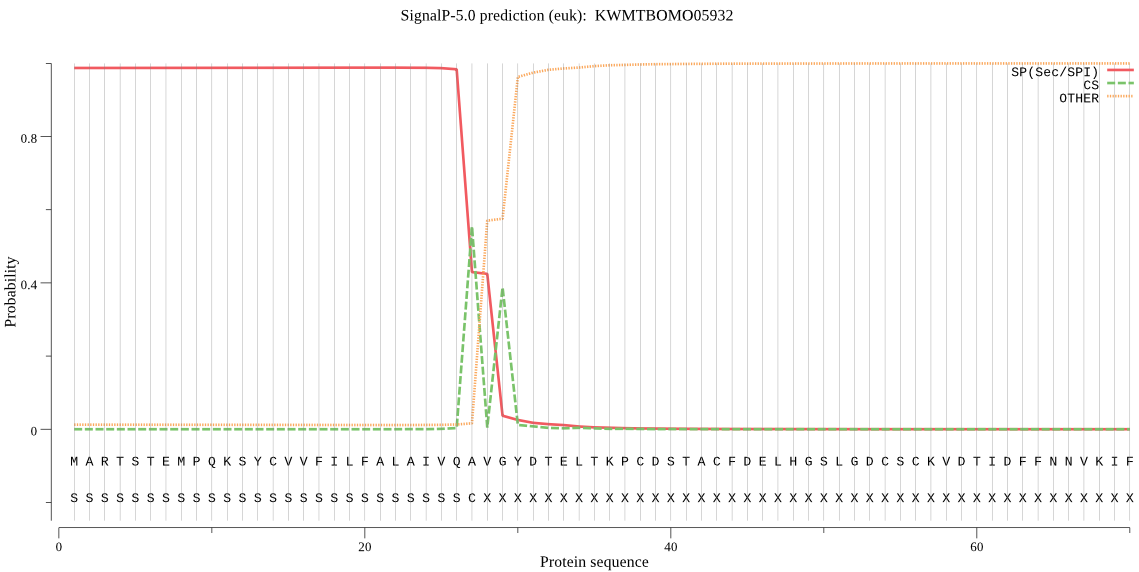
SignalP
Position: 1 - 27,
Likelihood: 0.988151
Length:
498
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.47324
Exp number, first 60 AAs:
18.47146
Total prob of N-in:
0.99425
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 30
outside
31 - 498

Population Genetic Test Statistics
Pi
262.535718
Theta
202.241178
Tajima's D
1.081951
CLR
0.090698
CSRT
0.686015699215039
Interpretation
Uncertain
