Gene
KWMTBOMO05931 Validated by peptides from experiments
Annotation
tetraspanin_E_[Bombyx_mori]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.566
Sequence
CDS
ATGAACTGGACCATGGTCAAAGACTTACTGAAGACCCATCTTGCTGTCGGTCCGTGGATCTTCATAGTCGTCGGGGCTGTAATGTTCGTGATCGCGTTCTTGGGATGCTGTGGAGCGATACGCGAGAGTCACTGCATGGTCGTCACGTACGCAATCTTCTTGCTGGTGATCATCATTGTGCAAGTGGTGATTAGCGTCCTCCTCTTCACCTACGGCGAGAGCATCAAGGAGAGCATTATGGATGGCGTCGGCGTGTTATTCAAAAAGAGATCTGACGCTAACGCAGACGAGGCCGCCGAGGCTGTCTTCTCTGAATTGCAACGACAGTTCGAATGCTGCGGTAACACTGGCGCTATCAACTACGGCCAGTTCACGTTGCCCGAGTCCTGTTGCGTCAAGAAGAGCATCCTCTCTACCTTCGCCGGCAACAACTGCACGGTCGACGCCGCGAATCCCGGCTGCGGTCCCAAGATCGGCGAGCTCTACCAGAAATGGAACAAGCCTATAGCCGGCGTTGCTCTCGGTGTTGCTTGTGTTGAGGTGGTAGGAGCCCTGTTCGCGCTGTGCCTCGCCAATTCCATAAGAAACATGGACAGAAGGTCACGCTACTAA
Protein
MNWTMVKDLLKTHLAVGPWIFIVVGAVMFVIAFLGCCGAIRESHCMVVTYAIFLLVIIIVQVVISVLLFTYGESIKESIMDGVGVLFKKRSDANADEAAEAVFSELQRQFECCGNTGAINYGQFTLPESCCVKKSILSTFAGNNCTVDAANPGCGPKIGELYQKWNKPIAGVALGVACVEVVGALFALCLANSIRNMDRRSRY
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Feature
chain Tetraspanin
Uniprot
Q2F647
Q2F648
A0A2A4IYD2
A0A194Q3K1
I4DJ82
I4DMY0
+ More
S4PWL6 A0A212EWY8 A0A1E1W4P8 A0A2H1WW71 A0A1E1WV10 Q9NB08 A0A0L7KT54 A0A0L7L5Y5 A0A194QWJ9 S4PF79 A0A1E1W0A4 A0A194RHI5 A0A194QGK8 A0A182ME04 I4DPV3 H9JFI9 A0A232FLJ2 K7IST2 E9J9S4 A0A1S4G0M8 Q16HX5 A0A224XJR8 A0A182VNU8 A0A212EGN7 A0A182TSU7 A0A1S4G0S2 A0A2A4K899 Q16HX6 A0A182XLJ0 A0A067QYE5 Q7PTA7 B0WVX8 A0A026WT07 A0A224XJF0 A0A3L8DMA2 T1E2B6 A0A2M4CTQ4 A0A2M4CV63 Q1HQ39 A0A182YML0 A0A1L8DKH3 A0A023EK84 A0A2M4BYW8 A0A182FGC4 A0A0C9Q5P6 A0A084WER4 A0A182PP40 A0A182KQN6 A0A1Q3FLS8 A0A023EK90 A0A182I2Q6 A7URL1 A0A182K3B6 A0A182IP56 A0A2M4A0S0 A0A182Q6M3 A0A3S2LT60 W5JA30 A0A2M3ZEZ2 A0A2M3YY18 A0A2M4CHV5 T1DQT0 T1GTV8 V5I9N6 N6U6Y0 A0A154PEQ3 A0A2M4A0K4 A0A1L8DJJ6 D6WZB8 A0A1Q3EUG6 A0A2M4CQ62 R4V3T2 A0A0J7L311 A0A2J7QP49 A0A2H1WF85 A0A2H1WFM4 A0A0L7R0A6 A0A182N923 T1I6I6 A0A182R3J5 R4G434 U5EYJ6 B4J3W7 A0A1S3FQI1 E2A972 E2B227 A0A0N7Z8R2 B3NAJ4 W8BV43 B4QDU2 B4HQF4 A0A076KUR5 J9K093 R4UW62
S4PWL6 A0A212EWY8 A0A1E1W4P8 A0A2H1WW71 A0A1E1WV10 Q9NB08 A0A0L7KT54 A0A0L7L5Y5 A0A194QWJ9 S4PF79 A0A1E1W0A4 A0A194RHI5 A0A194QGK8 A0A182ME04 I4DPV3 H9JFI9 A0A232FLJ2 K7IST2 E9J9S4 A0A1S4G0M8 Q16HX5 A0A224XJR8 A0A182VNU8 A0A212EGN7 A0A182TSU7 A0A1S4G0S2 A0A2A4K899 Q16HX6 A0A182XLJ0 A0A067QYE5 Q7PTA7 B0WVX8 A0A026WT07 A0A224XJF0 A0A3L8DMA2 T1E2B6 A0A2M4CTQ4 A0A2M4CV63 Q1HQ39 A0A182YML0 A0A1L8DKH3 A0A023EK84 A0A2M4BYW8 A0A182FGC4 A0A0C9Q5P6 A0A084WER4 A0A182PP40 A0A182KQN6 A0A1Q3FLS8 A0A023EK90 A0A182I2Q6 A7URL1 A0A182K3B6 A0A182IP56 A0A2M4A0S0 A0A182Q6M3 A0A3S2LT60 W5JA30 A0A2M3ZEZ2 A0A2M3YY18 A0A2M4CHV5 T1DQT0 T1GTV8 V5I9N6 N6U6Y0 A0A154PEQ3 A0A2M4A0K4 A0A1L8DJJ6 D6WZB8 A0A1Q3EUG6 A0A2M4CQ62 R4V3T2 A0A0J7L311 A0A2J7QP49 A0A2H1WF85 A0A2H1WFM4 A0A0L7R0A6 A0A182N923 T1I6I6 A0A182R3J5 R4G434 U5EYJ6 B4J3W7 A0A1S3FQI1 E2A972 E2B227 A0A0N7Z8R2 B3NAJ4 W8BV43 B4QDU2 B4HQF4 A0A076KUR5 J9K093 R4UW62
Pubmed
EMBL
DQ311225
ABD36170.1
DQ311224
ABD36169.1
NWSH01005565
PCG64123.1
+ More
KQ459472 KPJ00118.1 AK401350 BAM17972.1 AK402648 BAM19270.1 GAIX01006573 JAA85987.1 AGBW02011867 OWR45999.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 ODYU01011526 SOQ57310.1 GDQN01000423 JAT90631.1 AF274023 AAF90149.1 JTDY01006162 KOB66231.1 JTDY01002812 KOB70674.1 KQ461175 KPJ07946.1 GAIX01004167 JAA88393.1 GDQN01010636 JAT80418.1 KQ460154 KPJ17288.1 KQ458880 KPJ04637.1 AXCM01015250 AK403798 BAM19943.1 BABH01002470 BABH01002471 BABH01002472 NNAY01000043 OXU31611.1 AAZX01003309 AAZX01016448 GL769399 EFZ10437.1 CH478132 EAT33856.1 GFTR01003691 JAW12735.1 AGBW02015051 OWR40653.1 NWSH01000032 PCG80495.1 EAT33857.1 KK853109 KDR11258.1 AAAB01008807 EAA04202.4 DS232134 EDS35810.1 KK107128 EZA58234.1 GFTR01003819 JAW12607.1 QOIP01000006 RLU21537.1 GALA01000879 JAA93973.1 GGFL01004539 MBW68717.1 GGFL01004540 MBW68718.1 DQ443213 ABF51302.1 GFDF01007244 JAV06840.1 GAPW01004057 JAC09541.1 GGFJ01009053 MBW58194.1 GBYB01009418 JAG79185.1 ATLV01023246 KE525341 KFB48708.1 GFDL01006609 JAV28436.1 GAPW01004058 JAC09540.1 APCN01000178 EDO64653.1 GGFK01001038 MBW34359.1 AXCN02000540 RSAL01000264 RVE43294.1 ADMH02002070 ETN59619.1 GGFM01006249 MBW27000.1 GGFM01000400 MBW21151.1 GGFL01000705 MBW64883.1 GAMD01000985 JAB00606.1 CAQQ02086791 CAQQ02086792 GALX01002620 JAB65846.1 APGK01047299 APGK01047300 KB741077 KB631602 ENN74342.1 ERL84575.1 KQ434889 KZC10345.1 GGFK01001025 MBW34346.1 GFDF01007446 JAV06638.1 KQ971372 EFA09730.1 GFDL01016103 JAV18942.1 GGFL01003304 MBW67482.1 KC571964 AGM32463.1 LBMM01001031 KMQ97046.1 NEVH01012097 PNF30360.1 ODYU01008291 SOQ51745.1 ODYU01008292 SOQ51746.1 KQ414672 KOC64231.1 ACPB03011594 ACPB03011595 GAHY01001690 JAA75820.1 GANO01000315 JAB59556.1 CH916367 EDW01550.1 GL437711 EFN70057.1 GL445031 EFN60283.1 GDKW01002635 JAI53960.1 CH954177 EDV59748.1 GAMC01009385 GAMC01009384 JAB97171.1 CM000362 CM002911 EDX05954.1 KMY91897.1 CH480816 EDW46693.1 KF433416 AII97740.1 ABLF02025726 ABLF02025730 ABLF02025731 KC571982 AGM32481.1
KQ459472 KPJ00118.1 AK401350 BAM17972.1 AK402648 BAM19270.1 GAIX01006573 JAA85987.1 AGBW02011867 OWR45999.1 GDQN01010381 GDQN01009120 JAT80673.1 JAT81934.1 ODYU01011526 SOQ57310.1 GDQN01000423 JAT90631.1 AF274023 AAF90149.1 JTDY01006162 KOB66231.1 JTDY01002812 KOB70674.1 KQ461175 KPJ07946.1 GAIX01004167 JAA88393.1 GDQN01010636 JAT80418.1 KQ460154 KPJ17288.1 KQ458880 KPJ04637.1 AXCM01015250 AK403798 BAM19943.1 BABH01002470 BABH01002471 BABH01002472 NNAY01000043 OXU31611.1 AAZX01003309 AAZX01016448 GL769399 EFZ10437.1 CH478132 EAT33856.1 GFTR01003691 JAW12735.1 AGBW02015051 OWR40653.1 NWSH01000032 PCG80495.1 EAT33857.1 KK853109 KDR11258.1 AAAB01008807 EAA04202.4 DS232134 EDS35810.1 KK107128 EZA58234.1 GFTR01003819 JAW12607.1 QOIP01000006 RLU21537.1 GALA01000879 JAA93973.1 GGFL01004539 MBW68717.1 GGFL01004540 MBW68718.1 DQ443213 ABF51302.1 GFDF01007244 JAV06840.1 GAPW01004057 JAC09541.1 GGFJ01009053 MBW58194.1 GBYB01009418 JAG79185.1 ATLV01023246 KE525341 KFB48708.1 GFDL01006609 JAV28436.1 GAPW01004058 JAC09540.1 APCN01000178 EDO64653.1 GGFK01001038 MBW34359.1 AXCN02000540 RSAL01000264 RVE43294.1 ADMH02002070 ETN59619.1 GGFM01006249 MBW27000.1 GGFM01000400 MBW21151.1 GGFL01000705 MBW64883.1 GAMD01000985 JAB00606.1 CAQQ02086791 CAQQ02086792 GALX01002620 JAB65846.1 APGK01047299 APGK01047300 KB741077 KB631602 ENN74342.1 ERL84575.1 KQ434889 KZC10345.1 GGFK01001025 MBW34346.1 GFDF01007446 JAV06638.1 KQ971372 EFA09730.1 GFDL01016103 JAV18942.1 GGFL01003304 MBW67482.1 KC571964 AGM32463.1 LBMM01001031 KMQ97046.1 NEVH01012097 PNF30360.1 ODYU01008291 SOQ51745.1 ODYU01008292 SOQ51746.1 KQ414672 KOC64231.1 ACPB03011594 ACPB03011595 GAHY01001690 JAA75820.1 GANO01000315 JAB59556.1 CH916367 EDW01550.1 GL437711 EFN70057.1 GL445031 EFN60283.1 GDKW01002635 JAI53960.1 CH954177 EDV59748.1 GAMC01009385 GAMC01009384 JAB97171.1 CM000362 CM002911 EDX05954.1 KMY91897.1 CH480816 EDW46693.1 KF433416 AII97740.1 ABLF02025726 ABLF02025730 ABLF02025731 KC571982 AGM32481.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000053240
UP000075883
+ More
UP000005204 UP000215335 UP000002358 UP000008820 UP000075903 UP000075902 UP000076407 UP000027135 UP000007062 UP000002320 UP000053097 UP000279307 UP000076408 UP000069272 UP000030765 UP000075885 UP000075882 UP000075840 UP000075881 UP000075880 UP000075886 UP000283053 UP000000673 UP000015102 UP000019118 UP000030742 UP000076502 UP000007266 UP000036403 UP000235965 UP000053825 UP000075884 UP000015103 UP000075900 UP000001070 UP000081671 UP000000311 UP000008711 UP000000304 UP000001292 UP000007819
UP000005204 UP000215335 UP000002358 UP000008820 UP000075903 UP000075902 UP000076407 UP000027135 UP000007062 UP000002320 UP000053097 UP000279307 UP000076408 UP000069272 UP000030765 UP000075885 UP000075882 UP000075840 UP000075881 UP000075880 UP000075886 UP000283053 UP000000673 UP000015102 UP000019118 UP000030742 UP000076502 UP000007266 UP000036403 UP000235965 UP000053825 UP000075884 UP000015103 UP000075900 UP000001070 UP000081671 UP000000311 UP000008711 UP000000304 UP000001292 UP000007819
Pfam
PF00335 Tetraspanin
Interpro
Gene 3D
CDD
ProteinModelPortal
Q2F647
Q2F648
A0A2A4IYD2
A0A194Q3K1
I4DJ82
I4DMY0
+ More
S4PWL6 A0A212EWY8 A0A1E1W4P8 A0A2H1WW71 A0A1E1WV10 Q9NB08 A0A0L7KT54 A0A0L7L5Y5 A0A194QWJ9 S4PF79 A0A1E1W0A4 A0A194RHI5 A0A194QGK8 A0A182ME04 I4DPV3 H9JFI9 A0A232FLJ2 K7IST2 E9J9S4 A0A1S4G0M8 Q16HX5 A0A224XJR8 A0A182VNU8 A0A212EGN7 A0A182TSU7 A0A1S4G0S2 A0A2A4K899 Q16HX6 A0A182XLJ0 A0A067QYE5 Q7PTA7 B0WVX8 A0A026WT07 A0A224XJF0 A0A3L8DMA2 T1E2B6 A0A2M4CTQ4 A0A2M4CV63 Q1HQ39 A0A182YML0 A0A1L8DKH3 A0A023EK84 A0A2M4BYW8 A0A182FGC4 A0A0C9Q5P6 A0A084WER4 A0A182PP40 A0A182KQN6 A0A1Q3FLS8 A0A023EK90 A0A182I2Q6 A7URL1 A0A182K3B6 A0A182IP56 A0A2M4A0S0 A0A182Q6M3 A0A3S2LT60 W5JA30 A0A2M3ZEZ2 A0A2M3YY18 A0A2M4CHV5 T1DQT0 T1GTV8 V5I9N6 N6U6Y0 A0A154PEQ3 A0A2M4A0K4 A0A1L8DJJ6 D6WZB8 A0A1Q3EUG6 A0A2M4CQ62 R4V3T2 A0A0J7L311 A0A2J7QP49 A0A2H1WF85 A0A2H1WFM4 A0A0L7R0A6 A0A182N923 T1I6I6 A0A182R3J5 R4G434 U5EYJ6 B4J3W7 A0A1S3FQI1 E2A972 E2B227 A0A0N7Z8R2 B3NAJ4 W8BV43 B4QDU2 B4HQF4 A0A076KUR5 J9K093 R4UW62
S4PWL6 A0A212EWY8 A0A1E1W4P8 A0A2H1WW71 A0A1E1WV10 Q9NB08 A0A0L7KT54 A0A0L7L5Y5 A0A194QWJ9 S4PF79 A0A1E1W0A4 A0A194RHI5 A0A194QGK8 A0A182ME04 I4DPV3 H9JFI9 A0A232FLJ2 K7IST2 E9J9S4 A0A1S4G0M8 Q16HX5 A0A224XJR8 A0A182VNU8 A0A212EGN7 A0A182TSU7 A0A1S4G0S2 A0A2A4K899 Q16HX6 A0A182XLJ0 A0A067QYE5 Q7PTA7 B0WVX8 A0A026WT07 A0A224XJF0 A0A3L8DMA2 T1E2B6 A0A2M4CTQ4 A0A2M4CV63 Q1HQ39 A0A182YML0 A0A1L8DKH3 A0A023EK84 A0A2M4BYW8 A0A182FGC4 A0A0C9Q5P6 A0A084WER4 A0A182PP40 A0A182KQN6 A0A1Q3FLS8 A0A023EK90 A0A182I2Q6 A7URL1 A0A182K3B6 A0A182IP56 A0A2M4A0S0 A0A182Q6M3 A0A3S2LT60 W5JA30 A0A2M3ZEZ2 A0A2M3YY18 A0A2M4CHV5 T1DQT0 T1GTV8 V5I9N6 N6U6Y0 A0A154PEQ3 A0A2M4A0K4 A0A1L8DJJ6 D6WZB8 A0A1Q3EUG6 A0A2M4CQ62 R4V3T2 A0A0J7L311 A0A2J7QP49 A0A2H1WF85 A0A2H1WFM4 A0A0L7R0A6 A0A182N923 T1I6I6 A0A182R3J5 R4G434 U5EYJ6 B4J3W7 A0A1S3FQI1 E2A972 E2B227 A0A0N7Z8R2 B3NAJ4 W8BV43 B4QDU2 B4HQF4 A0A076KUR5 J9K093 R4UW62
PDB
5TCX
E-value=6.22621e-09,
Score=141
Ontologies
GO
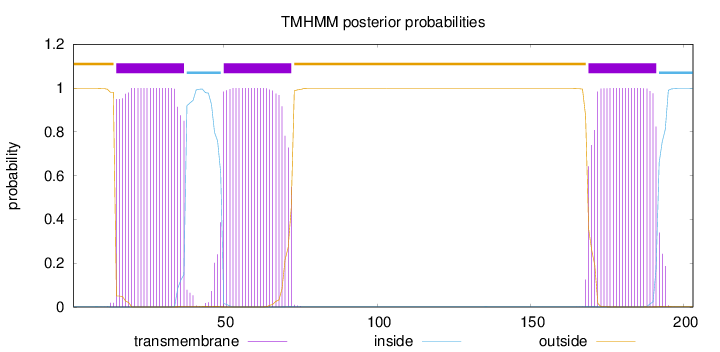
Topology
Subcellular location
Membrane
Length:
203
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
68.3625900000001
Exp number, first 60 AAs:
34.59575
Total prob of N-in:
0.00264
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 49
TMhelix
50 - 72
outside
73 - 168
TMhelix
169 - 191
inside
192 - 203
Population Genetic Test Statistics
Pi
207.13182
Theta
172.532797
Tajima's D
0.616034
CLR
1.214813
CSRT
0.546172691365432
Interpretation
Uncertain
