Pre Gene Modal
BGIBMGA002914
Annotation
adaptor_protein_complex-1_gamma_subunit_[Bombyx_mori]
Full name
AP-1 complex subunit gamma
Location in the cell
Cytoplasmic Reliability : 1.03 Golgi Reliability : 1.458
Sequence
CDS
ATGTACACTTTACTGCACGGCTTTTATAAGTATATAATACAGAAAGACGAATATTGCGTTCTAATACTCGGCCTAGACAATGCTGGCAAAACGACATATTTGGAAGCAACGAAAACAAAATTTACCAAAAAATATAAGGCTATGAACCCAAATAGGATAACAACAACCGTCGGACTTAATATAGGTAAAATAGATGTGAACGGTGTAAGACTTAATTTCTGGGACCTTGGTGGTCAGCAAGAATTACAATCCCTATGGGATAAGTACTATGCTGAATGCCATGCAGTAATCTACATAGTTGACTCATCAGATAGGGACAGGGTATCTGAATCCAAGGAAACCTTTGATAGAATGATATCCTCAGAACATTTAGCCGGTGTACCTTTGCTGGTGCTTGCGAACAAACAAGACATTCCAGACTGCATGGGCGTCCACACAGTGAAGCCCATATTCAATCAGAACGCACACCTGATTGGTGCTAGGGACATCATGTTAATGGCAACATCGGCATTAACTGGTGATGGCGTGGACGAAGGCATCCGATGGCTTGTAGACTGCATAAAACGCAACAGCGTCGATCGTCCTCCCCAGAAACAACTAGAAGAGTACGAGATGAACGGCTCCGAATCTGGGTTCAACCCGGCCTTCAACATTGCCACTATCAAGCAAGTGGTAAACGAGGCCATAGAGAGAGTGCGGATGCCGACACCGACCCGACTCAGGGATCTTATAAGGCAAATCAGAGCCGCTAGGACCGCCGCAGAAGAACGAAGCGTCGTCAACAAGGAATGCGCCTACATTAGGTCTACGTTCAGAGAAGAAGACAGCGTCTGGAGATGTCGCAACATTGCCAAGCTCCTGTACATCCACATGCTGGGTTATCCGGCCCATTTCGGTCAGCTGGAGTGCCTAAAGCTGATCGCAAGTCCTCGATTCACGGACAAACGAGTCGGCTACCTCGGCGCGATGCTGCTTCTGGACGAACGCCAAGATGTCCATCTCCTCATCACGAACTGCTTAAAGAACGACCTCAACAGCAACACACAGTTCGTGGTCGGGCTCGCGCTGTGCACCCTCGGAGCCATAGCCTCGCCGGAGATGGCCAGGGATTTGGCTTCGGAAGTGGAGCGTCTGATAAAGTCACCGAACGCCTACATAAAGAAGAAGGCGGCGCTTTGCGCGTTCAGGATCATCAGGAGAGTACCAGATTTAATGGAGATGTTCTTGCCGGCCACTAGAAGTCTACTTACCGAAAAGAATCATGGTGTCCTCATAACAGGCGTTACACTTATCACCGAAATGTGTGAGAACAGTCCCGACACGCTTAATCATTTCAAAAAGATCGTTCCTAATCTGGTGAGGATATTAAAGAATCTGATACTGGCCGGATATTCTCCGGAACATGACGTCAGCGGCGTATCCGATCCGTTCCTGCAGGTGAAGATACTGCGTCTGCTGCGGATACTGGGCAAGAACGACGCCGAAGCTTCGGAGGCGATGAACGACATATTGGCCCAGGTGGCCACCAACACGGAGACCAGCAAGAACGTCGGCAACACCATCCTGTACGAGACGGTGCTGTCGATAATGGACATCAAGTCCGAGAGCAGCCTCAGGGTGTTGGCCGTCAACATCCTGGGGAGGTTCCTGCTGAACAACGACAAGAACATCCGCTACGTCGCGCTCAACACGCTGCTGAGGACCGTGCACGTCGACACCTCGGCCGTGCAGAGGCACCGGACCACCATCCTCGAGTGTCTCAAGGACCCGGACATCTCGATCCGGCGACGCGCCATGGAGCTGTCGTTCGCGCTGGTGAACGGGCAGAACATCCGCGGCATGATGAAGGAGCTGCTGGCGTTCCTGGAGCGCAGCGACGCCGAGTTCAAGGCGCACTGCTCCAGCGCCGTGGTGCTCGCCGCCGAGCGGTACGCGCCCTCCGACAAGTGGCACCTCGACACGCTCTTCAAGGTCCTGCTCAAGGCTGGCAACTACTTACGAGACGACACGGTGTCGAGCACGATACAGATCATATCGTCAGCGGCGACGGAGCGTCAGGCGTACGGCGCCATGCGACTGTGGACTTCACTGGAACAGTCGGCTGTCTCGGGACTTGCCACAGAGAAACAACCTTTAATCCAGGTTGCCGCTTGGACTATTGGTGAATATGGAGATTTGTTAGTATCGGAAGCCAGCAGCGCTATCTCTATGGTGGATGAAGACGGTGTCGATGATTTTGTGCGACCGACTGAAGAATACGTGATTGACATATATCAGAAGCTGCTTTGGTCCACGCAGCTCTCGATCAGCACTAAAGAGTTCCTCTTACTTTCTTTGGCCAAACTGTCGACCAGATTCACCACGCAAGCCAGTCAGGAAAAAATAAGAGTGATCATAGACACGTTCGGATCTCACATCCACATCGAACTGCAGCAGCGAGGAGTTGAACTATCCCAACTCTTTAGACAATACGGGCACCTCCGACCCGCGCTGCTGGAGCGCATGCCCGCCATGGAGCCCGTCGCCGCGCCGCACCGCGACGAGCAGGAGCTCGATCTCAGCGACACCAGCCCTGACCACACTAAGCCTGACCAGGACGCCTTATTGGACTTGATCAACGGGTCTGATTCGTTGACGAACGGAGATGTGGAACACCAGCCTGAACAAATCGCTACCACCAACAATAATACTAGCAGTAATGACATCTTAGACTTGCTGAGCGGTCTAGATCTAACTCCGAGTGTACCCGCCGCCGCGCCCGCCGTGCTGCTCGCCAACAACAACGTGGAGCCGAGCGCCGCGCCCGCCGCGCTGCTGCTGGACGGGCTGTTCGCGCCGCCCGCGACCCCCCAACCCCCCCACCCGGCCGACGCTAATGAACGTAATGCGGCAGGGGCGGAGTGGGAGCGCGCGGTGCTGGAGCGCGGCGGCGTGCGCGTGTGCGTGTCGGCGCGGCGCGCGGCGGGCGCGGCGGCCACGCTCACGGCGCGCGCCACGTCGCAGCTGCACACGCTCACAGACTTCCTGTTCCAGGCCGCCGTGCCGAAGAAAATCCAGTTGGATATGATGTCTCCATCCGGTACGACGCTGTCGCCGCAGGGAGAGATCACGCAGGTGCTCAAAGTTACCAACCCGACCAAGACCCCGCTTCGGCTGCGGATAAGGGTGTCTTACAACATTGATGGCACCCCGGTATTGGAACAGACGGAGATCAACAATTTCCCGGCCGACCTGTTCAACTGA
Protein
MYTLLHGFYKYIIQKDEYCVLILGLDNAGKTTYLEATKTKFTKKYKAMNPNRITTTVGLNIGKIDVNGVRLNFWDLGGQQELQSLWDKYYAECHAVIYIVDSSDRDRVSESKETFDRMISSEHLAGVPLLVLANKQDIPDCMGVHTVKPIFNQNAHLIGARDIMLMATSALTGDGVDEGIRWLVDCIKRNSVDRPPQKQLEEYEMNGSESGFNPAFNIATIKQVVNEAIERVRMPTPTRLRDLIRQIRAARTAAEERSVVNKECAYIRSTFREEDSVWRCRNIAKLLYIHMLGYPAHFGQLECLKLIASPRFTDKRVGYLGAMLLLDERQDVHLLITNCLKNDLNSNTQFVVGLALCTLGAIASPEMARDLASEVERLIKSPNAYIKKKAALCAFRIIRRVPDLMEMFLPATRSLLTEKNHGVLITGVTLITEMCENSPDTLNHFKKIVPNLVRILKNLILAGYSPEHDVSGVSDPFLQVKILRLLRILGKNDAEASEAMNDILAQVATNTETSKNVGNTILYETVLSIMDIKSESSLRVLAVNILGRFLLNNDKNIRYVALNTLLRTVHVDTSAVQRHRTTILECLKDPDISIRRRAMELSFALVNGQNIRGMMKELLAFLERSDAEFKAHCSSAVVLAAERYAPSDKWHLDTLFKVLLKAGNYLRDDTVSSTIQIISSAATERQAYGAMRLWTSLEQSAVSGLATEKQPLIQVAAWTIGEYGDLLVSEASSAISMVDEDGVDDFVRPTEEYVIDIYQKLLWSTQLSISTKEFLLLSLAKLSTRFTTQASQEKIRVIIDTFGSHIHIELQQRGVELSQLFRQYGHLRPALLERMPAMEPVAAPHRDEQELDLSDTSPDHTKPDQDALLDLINGSDSLTNGDVEHQPEQIATTNNNTSSNDILDLLSGLDLTPSVPAAAPAVLLANNNVEPSAAPAALLLDGLFAPPATPQPPHPADANERNAAGAEWERAVLERGGVRVCVSARRAAGAAATLTARATSQLHTLTDFLFQAAVPKKIQLDMMSPSGTTLSPQGEITQVLKVTNPTKTPLRLRIRVSYNIDGTPVLEQTEINNFPADLFN
Summary
Similarity
Belongs to the adaptor complexes large subunit family.
Uniprot
EMBL
Proteomes
Interpro
IPR016024
ARM-type_fold
+ More
IPR008153 GAE_dom
IPR017107 AP1_complex_gsu
IPR013041 Clathrin_app_Ig-like_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR017104 AP2_complex_asu
IPR027417 P-loop_NTPase
IPR005225 Small_GTP-bd_dom
IPR006689 Small_GTPase_ARF/SAR
IPR036885 SWIB_MDM2_dom_sf
IPR019835 SWIB_domain
IPR003121 SWIB_MDM2_domain
IPR008153 GAE_dom
IPR017107 AP1_complex_gsu
IPR013041 Clathrin_app_Ig-like_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR017104 AP2_complex_asu
IPR027417 P-loop_NTPase
IPR005225 Small_GTP-bd_dom
IPR006689 Small_GTPase_ARF/SAR
IPR036885 SWIB_MDM2_dom_sf
IPR019835 SWIB_domain
IPR003121 SWIB_MDM2_domain
Gene 3D
ProteinModelPortal
PDB
6DFF
E-value=0,
Score=2070
Ontologies
GO
Topology
Subcellular location
Cytoplasm
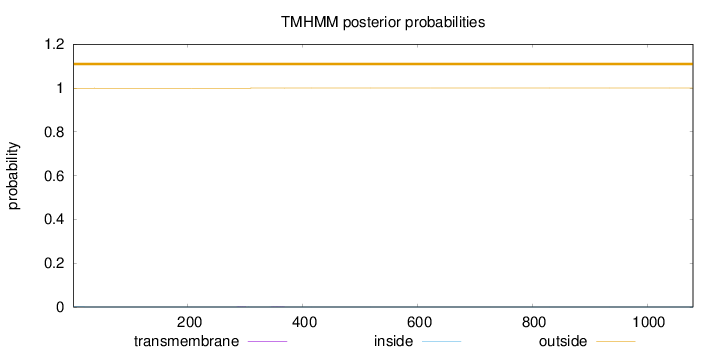
Length:
1080
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01446
Exp number, first 60 AAs:
0.00221
Total prob of N-in:
0.00064
outside
1 - 1080
Population Genetic Test Statistics
Pi
294.587319
Theta
184.302554
Tajima's D
1.727762
CLR
0.233528
CSRT
0.834908254587271
Interpretation
Uncertain
