Gene
KWMTBOMO05922
Pre Gene Modal
BGIBMGA002917
Annotation
protein_lin-7_homolog_C_[Papilio_xuthus]
Full name
Protein lin-7 homolog
Location in the cell
Cytoplasmic Reliability : 2.054 Mitochondrial Reliability : 2.073
Sequence
CDS
ATGACTACCACTGGCGAACAGTTAACGTTAGCTAGGGATATAAAACGTGCCATTGAATTACTCGAGAAACTGCAACAGAGTGGCGAAGTCCCTGCAACGAAATTAGCTGCTCTCCAGAAAGTTTTGCAGAGCGACTTTCTAAATGCCGTAAGAGAGGTTTACGAGCACGTCTACGAGACTGTAGACATACAAGGCTCCGCAGACATAAGAGCTTCCGCAACTGCAAAGGCAACGGTGGCCGCATTTGCTGCTGCAGAAGGACACGCTCACCCTCGAGTAGTGGAACTGCCGAAAACTGAGGAAGGCTTAGGGTTCAACGTAATGGGCGGAAAAGAACAAAATTCTCCGATTTATATATCTAGGATAATACCGGGCGGAGTTGCTGACAGACATGGTGGCTTGAAGAGGGGTGATCAACTTTTATCCGTTAACGGAGTCTCAGTTGAAGGCGAGAATCACGAGAAAGCCGTAGAACTTCTGAAGCAAGCCGTGGGATCCGTAAAATTAGTTGTCAGGTACACACCAAAGGTCTTGGAGGAAATGGAGATGAGATTCGACAAGCAAAGAACTACACGGCGACGCCAAAACTACAACTAA
Protein
MTTTGEQLTLARDIKRAIELLEKLQQSGEVPATKLAALQKVLQSDFLNAVREVYEHVYETVDIQGSADIRASATAKATVAAFAAAEGHAHPRVVELPKTEEGLGFNVMGGKEQNSPIYISRIIPGGVADRHGGLKRGDQLLSVNGVSVEGENHEKAVELLKQAVGSVKLVVRYTPKVLEEMEMRFDKQRTTRRRQNYN
Summary
Description
Plays a role in establishing and maintaining the asymmetric distribution of channels and receptors at the plasma membrane of polarized cells.
Similarity
Belongs to the lin-7 family.
Uniprot
H9J082
A0A3S2NF26
A0A2H1WWF0
A0A212F0B4
A0A194QRG8
I4DKU1
+ More
A0A1E1WAJ5 A0A194Q9L9 A0A0L7KWR0 A0A067RAA3 A0A0T6AWR3 A0A084WEA9 A0A1I8PFL9 T1PFI0 A0A182YEE2 A0A2M4AX28 A0A2M4C071 A0A182PTN3 A0A182LXS2 A0A182QQK0 A0A182S933 A0A182V8U0 A0A182NJD8 W5JA68 A0A182JRR7 A0A182WLB9 A0A182U5C1 A0A182XMC4 A0A182J880 Q7Q6G5 A0A182FH21 A0A182R4I3 A0A182I259 A0A023EL87 Q0IFH1 A0A0K8TQ47 A0A1A9UVH5 A0A1A9ZZ92 D3TKL0 A0A1W4XEJ4 T1DN00 A0A0A9XUG8 A0A1B6LZD5 A0A2R7VTL5 D6WII8 A0A1Q3FME3 B0WLT7 V5GZ23 A0A1A9YT36 A0A1B6DY19 A0A0V0G8J4 A0A069DX16 A0A1B6FIX6 B4JYU0 B4K4G9 A0A1D2NDN4 A0A0C9PS16 A0A034WI29 B4MCA0 W8C2S3 E0V9V6 A0A0K8UT62 A0A1J1J868 U5EU78 A0A0P4VVC7 R4FNR2 A0A336MF86 A0A1Y1L5P9 B3M1Y3 A0A195C3C9 A0A151J317 F4WK96 A0A151JWF6 A0A158NMM8 A0A151X0A2 A0A0A1WCY5 B4PUE7 B4IJE9 Q9VBY7 A0A023F9M1 E2BHA0 E2AW16 A0A026VY15 B3P6T1 A0A2A3E9Z2 A0A0M9A1V7 A0A088AVD7 A0A0L7R002 A0A1W4VKD7 B4NIX4 A0A2H8TLZ2 A0A226E756 A0A2S2NJD7 J9JW33 B4GE39 Q297G0 A0A224XXM3 A0A0C9PPT4 K7IVA8 K7INJ7
A0A1E1WAJ5 A0A194Q9L9 A0A0L7KWR0 A0A067RAA3 A0A0T6AWR3 A0A084WEA9 A0A1I8PFL9 T1PFI0 A0A182YEE2 A0A2M4AX28 A0A2M4C071 A0A182PTN3 A0A182LXS2 A0A182QQK0 A0A182S933 A0A182V8U0 A0A182NJD8 W5JA68 A0A182JRR7 A0A182WLB9 A0A182U5C1 A0A182XMC4 A0A182J880 Q7Q6G5 A0A182FH21 A0A182R4I3 A0A182I259 A0A023EL87 Q0IFH1 A0A0K8TQ47 A0A1A9UVH5 A0A1A9ZZ92 D3TKL0 A0A1W4XEJ4 T1DN00 A0A0A9XUG8 A0A1B6LZD5 A0A2R7VTL5 D6WII8 A0A1Q3FME3 B0WLT7 V5GZ23 A0A1A9YT36 A0A1B6DY19 A0A0V0G8J4 A0A069DX16 A0A1B6FIX6 B4JYU0 B4K4G9 A0A1D2NDN4 A0A0C9PS16 A0A034WI29 B4MCA0 W8C2S3 E0V9V6 A0A0K8UT62 A0A1J1J868 U5EU78 A0A0P4VVC7 R4FNR2 A0A336MF86 A0A1Y1L5P9 B3M1Y3 A0A195C3C9 A0A151J317 F4WK96 A0A151JWF6 A0A158NMM8 A0A151X0A2 A0A0A1WCY5 B4PUE7 B4IJE9 Q9VBY7 A0A023F9M1 E2BHA0 E2AW16 A0A026VY15 B3P6T1 A0A2A3E9Z2 A0A0M9A1V7 A0A088AVD7 A0A0L7R002 A0A1W4VKD7 B4NIX4 A0A2H8TLZ2 A0A226E756 A0A2S2NJD7 J9JW33 B4GE39 Q297G0 A0A224XXM3 A0A0C9PPT4 K7IVA8 K7INJ7
Pubmed
19121390
22118469
26354079
22651552
26227816
24845553
+ More
24438588 25315136 25244985 20920257 23761445 12364791 14747013 17210077 24945155 17510324 26369729 20353571 25401762 26823975 18362917 19820115 26334808 17994087 27289101 25348373 24495485 20566863 27129103 28004739 21719571 21347285 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 20798317 24508170 30249741 15632085 20075255
24438588 25315136 25244985 20920257 23761445 12364791 14747013 17210077 24945155 17510324 26369729 20353571 25401762 26823975 18362917 19820115 26334808 17994087 27289101 25348373 24495485 20566863 27129103 28004739 21719571 21347285 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25474469 20798317 24508170 30249741 15632085 20075255
EMBL
BABH01019079
RSAL01000147
RVE45862.1
ODYU01011572
SOQ57399.1
AGBW02011105
+ More
OWR47196.1 KQ461175 KPJ07934.1 AK401909 BAM18531.1 GDQN01007107 JAT83947.1 KQ459472 KPJ00106.1 JTDY01004985 KOB67506.1 KK852598 KDR20536.1 LJIG01022663 KRT79371.1 ATLV01023198 KE525341 KFB48553.1 KA647486 AFP62115.1 GGFK01011984 MBW45305.1 GGFJ01009579 MBW58720.1 AXCM01002346 AXCN02000448 ADMH02002052 GGFL01004222 ETN59745.1 MBW68400.1 AXCP01002184 AAAB01008960 EAA11636.3 APCN01000123 GAPW01004604 JAC08994.1 CH477317 EAT43743.1 GDAI01001583 JAI16020.1 EZ421942 EZ424293 ADD18238.1 ADD20569.1 GAMD01003290 JAA98300.1 GBHO01022764 GBHO01022731 GBRD01008851 GBRD01008850 GDHC01003417 JAG20840.1 JAG20873.1 JAG56970.1 JAQ15212.1 GEBQ01010977 JAT29000.1 KK854081 PTY10863.1 KQ971334 EEZ99659.1 GFDL01006351 JAV28694.1 DS231990 EDS30665.1 GALX01002748 JAB65718.1 GEDC01006769 JAS30529.1 GECL01002100 JAP04024.1 GBGD01003025 JAC85864.1 GECZ01019632 JAS50137.1 CH916378 EDV98555.1 CH933806 EDW13921.1 LJIJ01000076 ODN03370.1 GBYB01004158 JAG73925.1 GAKP01005164 GAKP01005162 GAKP01005161 JAC53790.1 CH940657 EDW63182.1 GAMC01000318 JAC06238.1 DS234996 EEB10162.1 GDHF01022563 JAI29751.1 CVRI01000075 CRL08576.1 GANO01002440 JAB57431.1 GDKW01000601 JAI55994.1 ACPB03023230 GAHY01001189 JAA76321.1 UFQT01000932 SSX28061.1 GEZM01063715 JAV69003.1 CH902617 EDV42243.1 KQ978350 KYM94683.1 KQ980313 KYN16720.1 GL888198 EGI65382.1 KQ981664 KYN38488.1 ADTU01002369 KQ982617 KYQ53786.1 GBXI01017545 JAC96746.1 CM000160 EDW98834.1 CH480848 EDW51140.1 AE014297 BT011021 AAF56389.1 AAR30181.1 GBBI01000622 JAC18090.1 GL448287 EFN84869.1 GL443213 EFN62438.1 KK107591 QOIP01000013 EZA48627.1 RLU15257.1 CH954182 EDV53751.1 KZ288311 PBC28545.1 KQ435762 KOX75507.1 KQ414673 KOC64142.1 CH964272 EDW84876.1 GFXV01003234 MBW15039.1 LNIX01000006 OXA52426.1 GGMR01004619 MBY17238.1 ABLF02023589 CH479182 EDW33874.1 CM000070 EAL28245.2 GFTR01003136 JAW13290.1 GBYB01003213 JAG72980.1 AAZX01001180
OWR47196.1 KQ461175 KPJ07934.1 AK401909 BAM18531.1 GDQN01007107 JAT83947.1 KQ459472 KPJ00106.1 JTDY01004985 KOB67506.1 KK852598 KDR20536.1 LJIG01022663 KRT79371.1 ATLV01023198 KE525341 KFB48553.1 KA647486 AFP62115.1 GGFK01011984 MBW45305.1 GGFJ01009579 MBW58720.1 AXCM01002346 AXCN02000448 ADMH02002052 GGFL01004222 ETN59745.1 MBW68400.1 AXCP01002184 AAAB01008960 EAA11636.3 APCN01000123 GAPW01004604 JAC08994.1 CH477317 EAT43743.1 GDAI01001583 JAI16020.1 EZ421942 EZ424293 ADD18238.1 ADD20569.1 GAMD01003290 JAA98300.1 GBHO01022764 GBHO01022731 GBRD01008851 GBRD01008850 GDHC01003417 JAG20840.1 JAG20873.1 JAG56970.1 JAQ15212.1 GEBQ01010977 JAT29000.1 KK854081 PTY10863.1 KQ971334 EEZ99659.1 GFDL01006351 JAV28694.1 DS231990 EDS30665.1 GALX01002748 JAB65718.1 GEDC01006769 JAS30529.1 GECL01002100 JAP04024.1 GBGD01003025 JAC85864.1 GECZ01019632 JAS50137.1 CH916378 EDV98555.1 CH933806 EDW13921.1 LJIJ01000076 ODN03370.1 GBYB01004158 JAG73925.1 GAKP01005164 GAKP01005162 GAKP01005161 JAC53790.1 CH940657 EDW63182.1 GAMC01000318 JAC06238.1 DS234996 EEB10162.1 GDHF01022563 JAI29751.1 CVRI01000075 CRL08576.1 GANO01002440 JAB57431.1 GDKW01000601 JAI55994.1 ACPB03023230 GAHY01001189 JAA76321.1 UFQT01000932 SSX28061.1 GEZM01063715 JAV69003.1 CH902617 EDV42243.1 KQ978350 KYM94683.1 KQ980313 KYN16720.1 GL888198 EGI65382.1 KQ981664 KYN38488.1 ADTU01002369 KQ982617 KYQ53786.1 GBXI01017545 JAC96746.1 CM000160 EDW98834.1 CH480848 EDW51140.1 AE014297 BT011021 AAF56389.1 AAR30181.1 GBBI01000622 JAC18090.1 GL448287 EFN84869.1 GL443213 EFN62438.1 KK107591 QOIP01000013 EZA48627.1 RLU15257.1 CH954182 EDV53751.1 KZ288311 PBC28545.1 KQ435762 KOX75507.1 KQ414673 KOC64142.1 CH964272 EDW84876.1 GFXV01003234 MBW15039.1 LNIX01000006 OXA52426.1 GGMR01004619 MBY17238.1 ABLF02023589 CH479182 EDW33874.1 CM000070 EAL28245.2 GFTR01003136 JAW13290.1 GBYB01003213 JAG72980.1 AAZX01001180
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000027135 UP000030765 UP000095300 UP000095301 UP000076408 UP000075885 UP000075883 UP000075886 UP000075901 UP000075903 UP000075884 UP000000673 UP000075881 UP000075920 UP000075902 UP000076407 UP000075880 UP000007062 UP000069272 UP000075900 UP000075840 UP000008820 UP000078200 UP000092445 UP000192223 UP000007266 UP000002320 UP000092443 UP000001070 UP000009192 UP000094527 UP000008792 UP000009046 UP000183832 UP000015103 UP000007801 UP000078542 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000002282 UP000001292 UP000000803 UP000008237 UP000000311 UP000053097 UP000279307 UP000008711 UP000242457 UP000053105 UP000005203 UP000053825 UP000192221 UP000007798 UP000198287 UP000007819 UP000008744 UP000001819 UP000002358
UP000027135 UP000030765 UP000095300 UP000095301 UP000076408 UP000075885 UP000075883 UP000075886 UP000075901 UP000075903 UP000075884 UP000000673 UP000075881 UP000075920 UP000075902 UP000076407 UP000075880 UP000007062 UP000069272 UP000075900 UP000075840 UP000008820 UP000078200 UP000092445 UP000192223 UP000007266 UP000002320 UP000092443 UP000001070 UP000009192 UP000094527 UP000008792 UP000009046 UP000183832 UP000015103 UP000007801 UP000078542 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000002282 UP000001292 UP000000803 UP000008237 UP000000311 UP000053097 UP000279307 UP000008711 UP000242457 UP000053105 UP000005203 UP000053825 UP000192221 UP000007798 UP000198287 UP000007819 UP000008744 UP000001819 UP000002358
Interpro
ProteinModelPortal
H9J082
A0A3S2NF26
A0A2H1WWF0
A0A212F0B4
A0A194QRG8
I4DKU1
+ More
A0A1E1WAJ5 A0A194Q9L9 A0A0L7KWR0 A0A067RAA3 A0A0T6AWR3 A0A084WEA9 A0A1I8PFL9 T1PFI0 A0A182YEE2 A0A2M4AX28 A0A2M4C071 A0A182PTN3 A0A182LXS2 A0A182QQK0 A0A182S933 A0A182V8U0 A0A182NJD8 W5JA68 A0A182JRR7 A0A182WLB9 A0A182U5C1 A0A182XMC4 A0A182J880 Q7Q6G5 A0A182FH21 A0A182R4I3 A0A182I259 A0A023EL87 Q0IFH1 A0A0K8TQ47 A0A1A9UVH5 A0A1A9ZZ92 D3TKL0 A0A1W4XEJ4 T1DN00 A0A0A9XUG8 A0A1B6LZD5 A0A2R7VTL5 D6WII8 A0A1Q3FME3 B0WLT7 V5GZ23 A0A1A9YT36 A0A1B6DY19 A0A0V0G8J4 A0A069DX16 A0A1B6FIX6 B4JYU0 B4K4G9 A0A1D2NDN4 A0A0C9PS16 A0A034WI29 B4MCA0 W8C2S3 E0V9V6 A0A0K8UT62 A0A1J1J868 U5EU78 A0A0P4VVC7 R4FNR2 A0A336MF86 A0A1Y1L5P9 B3M1Y3 A0A195C3C9 A0A151J317 F4WK96 A0A151JWF6 A0A158NMM8 A0A151X0A2 A0A0A1WCY5 B4PUE7 B4IJE9 Q9VBY7 A0A023F9M1 E2BHA0 E2AW16 A0A026VY15 B3P6T1 A0A2A3E9Z2 A0A0M9A1V7 A0A088AVD7 A0A0L7R002 A0A1W4VKD7 B4NIX4 A0A2H8TLZ2 A0A226E756 A0A2S2NJD7 J9JW33 B4GE39 Q297G0 A0A224XXM3 A0A0C9PPT4 K7IVA8 K7INJ7
A0A1E1WAJ5 A0A194Q9L9 A0A0L7KWR0 A0A067RAA3 A0A0T6AWR3 A0A084WEA9 A0A1I8PFL9 T1PFI0 A0A182YEE2 A0A2M4AX28 A0A2M4C071 A0A182PTN3 A0A182LXS2 A0A182QQK0 A0A182S933 A0A182V8U0 A0A182NJD8 W5JA68 A0A182JRR7 A0A182WLB9 A0A182U5C1 A0A182XMC4 A0A182J880 Q7Q6G5 A0A182FH21 A0A182R4I3 A0A182I259 A0A023EL87 Q0IFH1 A0A0K8TQ47 A0A1A9UVH5 A0A1A9ZZ92 D3TKL0 A0A1W4XEJ4 T1DN00 A0A0A9XUG8 A0A1B6LZD5 A0A2R7VTL5 D6WII8 A0A1Q3FME3 B0WLT7 V5GZ23 A0A1A9YT36 A0A1B6DY19 A0A0V0G8J4 A0A069DX16 A0A1B6FIX6 B4JYU0 B4K4G9 A0A1D2NDN4 A0A0C9PS16 A0A034WI29 B4MCA0 W8C2S3 E0V9V6 A0A0K8UT62 A0A1J1J868 U5EU78 A0A0P4VVC7 R4FNR2 A0A336MF86 A0A1Y1L5P9 B3M1Y3 A0A195C3C9 A0A151J317 F4WK96 A0A151JWF6 A0A158NMM8 A0A151X0A2 A0A0A1WCY5 B4PUE7 B4IJE9 Q9VBY7 A0A023F9M1 E2BHA0 E2AW16 A0A026VY15 B3P6T1 A0A2A3E9Z2 A0A0M9A1V7 A0A088AVD7 A0A0L7R002 A0A1W4VKD7 B4NIX4 A0A2H8TLZ2 A0A226E756 A0A2S2NJD7 J9JW33 B4GE39 Q297G0 A0A224XXM3 A0A0C9PPT4 K7IVA8 K7INJ7
PDB
2DKR
E-value=1.78082e-34,
Score=361
Ontologies
GO
Topology
Subcellular location
Cell membrane
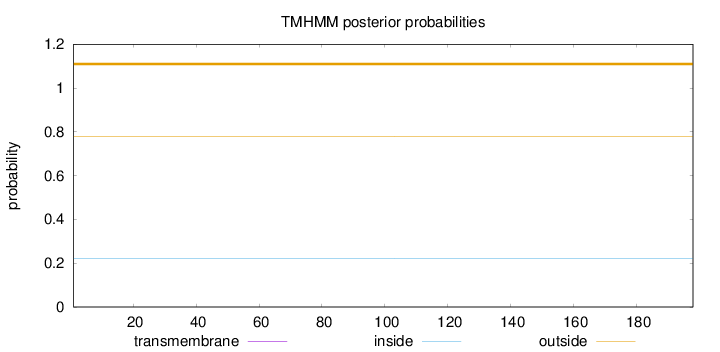
Length:
198
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000490000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22254
outside
1 - 198
Population Genetic Test Statistics
Pi
119.267875
Theta
92.52601
Tajima's D
0.528386
CLR
34.095772
CSRT
0.51812409379531
Interpretation
Uncertain
