Gene
KWMTBOMO05921
Pre Gene Modal
BGIBMGA002786
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase-like_2_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.424
Sequence
CDS
ATGGGGAAACGACAGCATCAAAAAGATAAAATGTATTTAACGTATACGGAATGGACAACTCTCTACGGTGGAAAACGTTCTGGAACGGCTACTGAAGAAGATACAACATTTAAAAGACTTCCATTCGATCACTGTTGTCTTTGTTTGCAGCCCTTTGACGATCCCTACTGTGATGCAGACGGCAATGTATTTGAATTGCAAGCTATCATTGATTTTCGAAAAAAGTTTAAAATTAATCCTGTGACTGGAAAGAAACTTGATATAAAAACTCTAATAAAGTTAAACTTCTTCAAAAATGCTGAAGATGCCTACCACTGCCCAGTATTATTTAAACCCTTTACAAAAAACTCGCATATTGTTGCCATACGCACTAGTGGAAATGTCTACTCTTATGAGGCTGTTGAACAGCTCAATATCAAAGGCAAAAATTGGAAGGACCTGGTTGATGATACACCCTTTGCTGCACGTAATGATATTATCACTATTCAGGACCCTCAAAACTTGGCAAAGTTTAATATATCAAATTTTCATCACATAAAACACAATTTACGTGTTGAAACTGAAGAGGAAATAGCATTGAAAAAAGACCCACATGCAAGATTGAAAACAGTATCTGCCGAAACAAAAGATATATTACAAGAATTAGAAAAGGACTATAAGGCACCAGAAAAGAAAGAAGATCAAAAAGAGTCTGCCGACAAATTCAATGCGGCCCACTATTCCACTGGGAAGGTCGCGGCCAGCTTCACATCGACAGCAATGGTACCGGAGACGACACACGAAGCAGCCATCATATGTGAAGATGAGGTTATTTATGAAAGAACAAAAAAGAAAGGTTATGTAAGACTGGTAACAAATGTAGGACATCTGAACTTTGAATTGTACTGCGACGTAACGCCCAAAACTTGTCACAACTTCATGACACATTGTAAAAATGGATACTACAACGGAACAAAGTTCCACAGGTCTATCAGGAATTTTATGATTCAAGGAGGAGATCCAACGGGCACAGGGTTGGGAGGCGAATCCATTTGGAAGAAACCTTTTGATGATGAAATTAAGCCAAACTTGCACCACACCGGACGAGGAGTCTTGTCTATGGCTAACTCGGGAACCAATACTAACGGATCACAATTTTTCATTACGTTCCGTTCGTGCAAACAGTTGGACGGTAAACACACTATATTCGGGAAGCTGGTCGGCGGTCTGGACACTTTGAACGCCATGGAGTCGATTGAGGTCGACAATAAAGACAGGCCCATACAGGATATAGTTATTGAGGCAGCGCAAGTCTTTGTGGATCCTTTCGCTGAGGCTGAAGAGGAGCTCGCTAAAGAACGGGCAGCAGAATTAAAGCGTAAAGCTGAAGAGAAGGATCCCGGAGTCAGACCTAAAAAAGCGACACAACCCCTTAAAGTATTCAGGGAAGGTGTCGGTAAATACTTGAACCTGCAAGAGTCTGCAACGAAACCTGGAAAAGACCAGGAAATTCCAGCAAAAAAACCCAAAAAAGATGTCACTAGTTTTGGTAATTTTGATACCTGGTAA
Protein
MGKRQHQKDKMYLTYTEWTTLYGGKRSGTATEEDTTFKRLPFDHCCLCLQPFDDPYCDADGNVFELQAIIDFRKKFKINPVTGKKLDIKTLIKLNFFKNAEDAYHCPVLFKPFTKNSHIVAIRTSGNVYSYEAVEQLNIKGKNWKDLVDDTPFAARNDIITIQDPQNLAKFNISNFHHIKHNLRVETEEEIALKKDPHARLKTVSAETKDILQELEKDYKAPEKKEDQKESADKFNAAHYSTGKVAASFTSTAMVPETTHEAAIICEDEVIYERTKKKGYVRLVTNVGHLNFELYCDVTPKTCHNFMTHCKNGYYNGTKFHRSIRNFMIQGGDPTGTGLGGESIWKKPFDDEIKPNLHHTGRGVLSMANSGTNTNGSQFFITFRSCKQLDGKHTIFGKLVGGLDTLNAMESIEVDNKDRPIQDIVIEAAQVFVDPFAEAEEELAKERAAELKRKAEEKDPGVRPKKATQPLKVFREGVGKYLNLQESATKPGKDQEIPAKKPKKDVTSFGNFDTW
Summary
Uniprot
H9IZV1
A0A3S2NVS1
A0A212F0G0
A0A194Q526
S4PE85
A0A2A4IYS6
+ More
A0A2H1WWG3 A0A0L7KWK8 A0A1B0CIX4 Q16NE3 Q16IE1 A0A1B6F3U2 A0A2C9GQV5 A0A182KSY6 A0A0L0BZX7 A0A182V0Z0 A0A084WHF9 A0A182WZX7 A0A182K8H1 A0A182UH27 A0A1B6I137 A0A182W9D7 B0WTD2 A0A182Y216 A0A034VCZ2 A0A182RQV9 A0A182P1S2 A0A182JKM0 A0A1Q3FCS0 A0A026WII7 A0A1I8MD16 A0A0K8UBJ6 A0A151WR25 A0A1L8DUJ5 A0A182N2L6 A0A1B0DP53 A0A182M705 B3MFM6 W8AVZ5 A0A182QN32 A0A195EH79 A0A1I8P751 W5JD36 F4WPR2 A0A1A9X4W7 A0A182FZR8 A0A154P3D5 K7IWT7 E2APB7 A0A158NHX7 A0A1W4UKZ6 E9IBN0 B4HSV9 A0A1Y1MUX8 A0A1B6F8R0 A0A232EZG8 B3NPK8 A0A336M6W0 A0A1B6DSR5 A0A195C087 A0A0L7R6Z9 A0A0M4E7A1 A0A336MC90 A0A1L8DU38 A0A2A3E544 A0A1B0AF55 A0A0J9RDU0 Q7K231 B4KNZ0 A0A3B0J2Q3 B4GAC7 A0A1A9YFZ5 A0A1B0BS11 B4MJX8 A0A067QFD2 B4P694 Q291S0 A0A0C9QRT2 A0A088A006 A0A0K8TKP6 A0A2J7QZJ1 A0A1A9VU64 A0A0J7KXW6 A0A1B0FPY4 A0A195AWC9 B4LIP4 E0VTT9 A0A195FFJ3 A0A0C9R0H6 B4J466 A0A1B6CWT2 A0A2J7QZJ7 N6USE6 A0A0N0BDW3 D6X1Y7 A0A1W4XIZ5 E2C8K5 U4TY59 A0A0P5V612 E9H7X4
A0A2H1WWG3 A0A0L7KWK8 A0A1B0CIX4 Q16NE3 Q16IE1 A0A1B6F3U2 A0A2C9GQV5 A0A182KSY6 A0A0L0BZX7 A0A182V0Z0 A0A084WHF9 A0A182WZX7 A0A182K8H1 A0A182UH27 A0A1B6I137 A0A182W9D7 B0WTD2 A0A182Y216 A0A034VCZ2 A0A182RQV9 A0A182P1S2 A0A182JKM0 A0A1Q3FCS0 A0A026WII7 A0A1I8MD16 A0A0K8UBJ6 A0A151WR25 A0A1L8DUJ5 A0A182N2L6 A0A1B0DP53 A0A182M705 B3MFM6 W8AVZ5 A0A182QN32 A0A195EH79 A0A1I8P751 W5JD36 F4WPR2 A0A1A9X4W7 A0A182FZR8 A0A154P3D5 K7IWT7 E2APB7 A0A158NHX7 A0A1W4UKZ6 E9IBN0 B4HSV9 A0A1Y1MUX8 A0A1B6F8R0 A0A232EZG8 B3NPK8 A0A336M6W0 A0A1B6DSR5 A0A195C087 A0A0L7R6Z9 A0A0M4E7A1 A0A336MC90 A0A1L8DU38 A0A2A3E544 A0A1B0AF55 A0A0J9RDU0 Q7K231 B4KNZ0 A0A3B0J2Q3 B4GAC7 A0A1A9YFZ5 A0A1B0BS11 B4MJX8 A0A067QFD2 B4P694 Q291S0 A0A0C9QRT2 A0A088A006 A0A0K8TKP6 A0A2J7QZJ1 A0A1A9VU64 A0A0J7KXW6 A0A1B0FPY4 A0A195AWC9 B4LIP4 E0VTT9 A0A195FFJ3 A0A0C9R0H6 B4J466 A0A1B6CWT2 A0A2J7QZJ7 N6USE6 A0A0N0BDW3 D6X1Y7 A0A1W4XIZ5 E2C8K5 U4TY59 A0A0P5V612 E9H7X4
Pubmed
19121390
22118469
26354079
23622113
26227816
17510324
+ More
20966253 26108605 24438588 25244985 25348373 24508170 30249741 25315136 17994087 24495485 20920257 23761445 21719571 20075255 20798317 21347285 21282665 28004739 28648823 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 17550304 15632085 26369729 20566863 23537049 18362917 19820115 21292972
20966253 26108605 24438588 25244985 25348373 24508170 30249741 25315136 17994087 24495485 20920257 23761445 21719571 20075255 20798317 21347285 21282665 28004739 28648823 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 17550304 15632085 26369729 20566863 23537049 18362917 19820115 21292972
EMBL
BABH01019079
RSAL01000147
RVE45863.1
AGBW02011105
OWR47197.1
KQ459472
+ More
KPJ00105.1 GAIX01004642 JAA87918.1 NWSH01004572 PCG64911.1 ODYU01011572 SOQ57398.1 JTDY01004985 KOB67505.1 AJWK01013857 CH477828 EAT35856.1 CH478086 EAT34025.1 GECZ01024889 JAS44880.1 APCN01003275 JRES01001096 KNC25605.1 ATLV01023810 ATLV01023811 ATLV01023812 KE525347 KFB49653.1 GECU01027074 JAS80632.1 DS232085 EDS34370.1 GAKP01017791 JAC41161.1 GFDL01009670 JAV25375.1 KK107185 QOIP01000012 EZA55830.1 RLU16370.1 GDHF01028373 JAI23941.1 KQ982813 KYQ50316.1 GFDF01004149 JAV09935.1 AJVK01018086 AXCM01009508 CH902619 EDV37716.1 GAMC01017647 JAB88908.1 AXCN02001078 KQ978957 KYN27219.1 ADMH02001585 ETN61966.1 GL888255 EGI63803.1 KQ434809 KZC06381.1 AAZX01011511 GL441504 EFN64716.1 ADTU01016101 GL762111 EFZ22150.1 CH480816 EDW48123.1 GEZM01020010 JAV89461.1 GECZ01023151 JAS46618.1 NNAY01001516 OXU23719.1 CH954179 EDV55775.1 UFQT01000640 SSX26006.1 GEDC01008595 JAS28703.1 KQ978501 KYM93563.1 KQ414643 KOC66650.1 CP012524 ALC42528.1 SSX26007.1 GFDF01004160 JAV09924.1 KZ288379 PBC26614.1 CM002911 KMY94238.1 AE013599 AY061295 AAF58019.1 AAL28843.1 CH933808 EDW09035.1 OUUW01000001 SPP73422.1 CH479181 EDW31879.1 JXJN01019442 CH963846 EDW72417.1 KK853603 KDR06370.1 CM000158 EDW91944.1 CM000071 EAL25042.2 GBYB01006439 JAG76206.1 GDAI01002902 JAI14701.1 NEVH01009069 PNF33993.1 LBMM01002197 KMQ95156.1 CCAG010004164 KQ976725 KYM76553.1 CH940648 EDW61397.1 DS235773 EEB16795.1 KQ981636 KYN38794.1 GBYB01006437 JAG76204.1 CH916367 EDW02672.1 GEDC01019655 JAS17643.1 PNF33994.1 APGK01018457 APGK01018458 APGK01018459 KB740076 ENN81647.1 KQ435848 KOX71004.1 KQ971371 EFA10186.1 GL453666 EFN75735.1 KB631643 ERL84928.1 GDIP01222092 GDIP01119826 GDIP01089713 GDIP01081402 LRGB01003325 JAL83888.1 KZS03253.1 GL732602 EFX72103.1
KPJ00105.1 GAIX01004642 JAA87918.1 NWSH01004572 PCG64911.1 ODYU01011572 SOQ57398.1 JTDY01004985 KOB67505.1 AJWK01013857 CH477828 EAT35856.1 CH478086 EAT34025.1 GECZ01024889 JAS44880.1 APCN01003275 JRES01001096 KNC25605.1 ATLV01023810 ATLV01023811 ATLV01023812 KE525347 KFB49653.1 GECU01027074 JAS80632.1 DS232085 EDS34370.1 GAKP01017791 JAC41161.1 GFDL01009670 JAV25375.1 KK107185 QOIP01000012 EZA55830.1 RLU16370.1 GDHF01028373 JAI23941.1 KQ982813 KYQ50316.1 GFDF01004149 JAV09935.1 AJVK01018086 AXCM01009508 CH902619 EDV37716.1 GAMC01017647 JAB88908.1 AXCN02001078 KQ978957 KYN27219.1 ADMH02001585 ETN61966.1 GL888255 EGI63803.1 KQ434809 KZC06381.1 AAZX01011511 GL441504 EFN64716.1 ADTU01016101 GL762111 EFZ22150.1 CH480816 EDW48123.1 GEZM01020010 JAV89461.1 GECZ01023151 JAS46618.1 NNAY01001516 OXU23719.1 CH954179 EDV55775.1 UFQT01000640 SSX26006.1 GEDC01008595 JAS28703.1 KQ978501 KYM93563.1 KQ414643 KOC66650.1 CP012524 ALC42528.1 SSX26007.1 GFDF01004160 JAV09924.1 KZ288379 PBC26614.1 CM002911 KMY94238.1 AE013599 AY061295 AAF58019.1 AAL28843.1 CH933808 EDW09035.1 OUUW01000001 SPP73422.1 CH479181 EDW31879.1 JXJN01019442 CH963846 EDW72417.1 KK853603 KDR06370.1 CM000158 EDW91944.1 CM000071 EAL25042.2 GBYB01006439 JAG76206.1 GDAI01002902 JAI14701.1 NEVH01009069 PNF33993.1 LBMM01002197 KMQ95156.1 CCAG010004164 KQ976725 KYM76553.1 CH940648 EDW61397.1 DS235773 EEB16795.1 KQ981636 KYN38794.1 GBYB01006437 JAG76204.1 CH916367 EDW02672.1 GEDC01019655 JAS17643.1 PNF33994.1 APGK01018457 APGK01018458 APGK01018459 KB740076 ENN81647.1 KQ435848 KOX71004.1 KQ971371 EFA10186.1 GL453666 EFN75735.1 KB631643 ERL84928.1 GDIP01222092 GDIP01119826 GDIP01089713 GDIP01081402 LRGB01003325 JAL83888.1 KZS03253.1 GL732602 EFX72103.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000037510
+ More
UP000092461 UP000008820 UP000075840 UP000075882 UP000037069 UP000075903 UP000030765 UP000076407 UP000075881 UP000075902 UP000075920 UP000002320 UP000076408 UP000075900 UP000075885 UP000075880 UP000053097 UP000279307 UP000095301 UP000075809 UP000075884 UP000092462 UP000075883 UP000007801 UP000075886 UP000078492 UP000095300 UP000000673 UP000007755 UP000091820 UP000069272 UP000076502 UP000002358 UP000000311 UP000005205 UP000192221 UP000001292 UP000215335 UP000008711 UP000078542 UP000053825 UP000092553 UP000242457 UP000092445 UP000000803 UP000009192 UP000268350 UP000008744 UP000092443 UP000092460 UP000007798 UP000027135 UP000002282 UP000001819 UP000005203 UP000235965 UP000078200 UP000036403 UP000092444 UP000078540 UP000008792 UP000009046 UP000078541 UP000001070 UP000019118 UP000053105 UP000007266 UP000192223 UP000008237 UP000030742 UP000076858 UP000000305
UP000092461 UP000008820 UP000075840 UP000075882 UP000037069 UP000075903 UP000030765 UP000076407 UP000075881 UP000075902 UP000075920 UP000002320 UP000076408 UP000075900 UP000075885 UP000075880 UP000053097 UP000279307 UP000095301 UP000075809 UP000075884 UP000092462 UP000075883 UP000007801 UP000075886 UP000078492 UP000095300 UP000000673 UP000007755 UP000091820 UP000069272 UP000076502 UP000002358 UP000000311 UP000005205 UP000192221 UP000001292 UP000215335 UP000008711 UP000078542 UP000053825 UP000092553 UP000242457 UP000092445 UP000000803 UP000009192 UP000268350 UP000008744 UP000092443 UP000092460 UP000007798 UP000027135 UP000002282 UP000001819 UP000005203 UP000235965 UP000078200 UP000036403 UP000092444 UP000078540 UP000008792 UP000009046 UP000078541 UP000001070 UP000019118 UP000053105 UP000007266 UP000192223 UP000008237 UP000030742 UP000076858 UP000000305
PRIDE
Interpro
SUPFAM
SSF50891
SSF50891
Gene 3D
ProteinModelPortal
H9IZV1
A0A3S2NVS1
A0A212F0G0
A0A194Q526
S4PE85
A0A2A4IYS6
+ More
A0A2H1WWG3 A0A0L7KWK8 A0A1B0CIX4 Q16NE3 Q16IE1 A0A1B6F3U2 A0A2C9GQV5 A0A182KSY6 A0A0L0BZX7 A0A182V0Z0 A0A084WHF9 A0A182WZX7 A0A182K8H1 A0A182UH27 A0A1B6I137 A0A182W9D7 B0WTD2 A0A182Y216 A0A034VCZ2 A0A182RQV9 A0A182P1S2 A0A182JKM0 A0A1Q3FCS0 A0A026WII7 A0A1I8MD16 A0A0K8UBJ6 A0A151WR25 A0A1L8DUJ5 A0A182N2L6 A0A1B0DP53 A0A182M705 B3MFM6 W8AVZ5 A0A182QN32 A0A195EH79 A0A1I8P751 W5JD36 F4WPR2 A0A1A9X4W7 A0A182FZR8 A0A154P3D5 K7IWT7 E2APB7 A0A158NHX7 A0A1W4UKZ6 E9IBN0 B4HSV9 A0A1Y1MUX8 A0A1B6F8R0 A0A232EZG8 B3NPK8 A0A336M6W0 A0A1B6DSR5 A0A195C087 A0A0L7R6Z9 A0A0M4E7A1 A0A336MC90 A0A1L8DU38 A0A2A3E544 A0A1B0AF55 A0A0J9RDU0 Q7K231 B4KNZ0 A0A3B0J2Q3 B4GAC7 A0A1A9YFZ5 A0A1B0BS11 B4MJX8 A0A067QFD2 B4P694 Q291S0 A0A0C9QRT2 A0A088A006 A0A0K8TKP6 A0A2J7QZJ1 A0A1A9VU64 A0A0J7KXW6 A0A1B0FPY4 A0A195AWC9 B4LIP4 E0VTT9 A0A195FFJ3 A0A0C9R0H6 B4J466 A0A1B6CWT2 A0A2J7QZJ7 N6USE6 A0A0N0BDW3 D6X1Y7 A0A1W4XIZ5 E2C8K5 U4TY59 A0A0P5V612 E9H7X4
A0A2H1WWG3 A0A0L7KWK8 A0A1B0CIX4 Q16NE3 Q16IE1 A0A1B6F3U2 A0A2C9GQV5 A0A182KSY6 A0A0L0BZX7 A0A182V0Z0 A0A084WHF9 A0A182WZX7 A0A182K8H1 A0A182UH27 A0A1B6I137 A0A182W9D7 B0WTD2 A0A182Y216 A0A034VCZ2 A0A182RQV9 A0A182P1S2 A0A182JKM0 A0A1Q3FCS0 A0A026WII7 A0A1I8MD16 A0A0K8UBJ6 A0A151WR25 A0A1L8DUJ5 A0A182N2L6 A0A1B0DP53 A0A182M705 B3MFM6 W8AVZ5 A0A182QN32 A0A195EH79 A0A1I8P751 W5JD36 F4WPR2 A0A1A9X4W7 A0A182FZR8 A0A154P3D5 K7IWT7 E2APB7 A0A158NHX7 A0A1W4UKZ6 E9IBN0 B4HSV9 A0A1Y1MUX8 A0A1B6F8R0 A0A232EZG8 B3NPK8 A0A336M6W0 A0A1B6DSR5 A0A195C087 A0A0L7R6Z9 A0A0M4E7A1 A0A336MC90 A0A1L8DU38 A0A2A3E544 A0A1B0AF55 A0A0J9RDU0 Q7K231 B4KNZ0 A0A3B0J2Q3 B4GAC7 A0A1A9YFZ5 A0A1B0BS11 B4MJX8 A0A067QFD2 B4P694 Q291S0 A0A0C9QRT2 A0A088A006 A0A0K8TKP6 A0A2J7QZJ1 A0A1A9VU64 A0A0J7KXW6 A0A1B0FPY4 A0A195AWC9 B4LIP4 E0VTT9 A0A195FFJ3 A0A0C9R0H6 B4J466 A0A1B6CWT2 A0A2J7QZJ7 N6USE6 A0A0N0BDW3 D6X1Y7 A0A1W4XIZ5 E2C8K5 U4TY59 A0A0P5V612 E9H7X4
PDB
1ZKC
E-value=2.34189e-54,
Score=538
Ontologies
GO
Topology
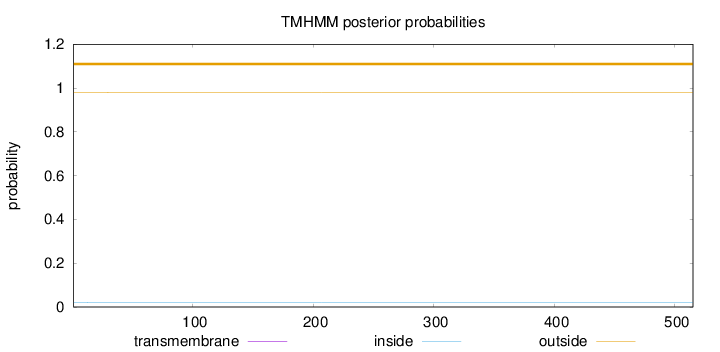
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00405000000000001
Exp number, first 60 AAs:
0.0015
Total prob of N-in:
0.01980
outside
1 - 515
Population Genetic Test Statistics
Pi
175.20392
Theta
143.08982
Tajima's D
-1.198556
CLR
134.014614
CSRT
0.103794810259487
Interpretation
Uncertain
