Gene
KWMTBOMO05919
Annotation
tetraspanin_D107_[Bombyx_mori]
Full name
Tetraspanin
Location in the cell
Extracellular Reliability : 3.097
Sequence
CDS
ATGTTTTCAGTATTCCTCTTGATCATATTCGTGGCTGAACTGGCTGTGGGCATCGCTGGTTACATGAAGCATACGGACTTGGAAGATTCGGTTATGCGCAACCTGAACGCTTCCATTACTCAATATCCGGTAGACAAGAACGTCCAGAAGACTATTGATATCATACAAACAGACTTGCAATGCTGCGGTATAAACAGTCCGGCCGACTGGGCCGATCACGGTCTGCCCATACCGAGCACGTGCTGCTCCGCCCAGGAGATCAATGACGGCGTCGTCGCGGCCTGCACCGAGAACTCGACCAACTTCCACTCGAAGGGATGTCTTACTAAACTGGTTGTTCACATGAAGGACATTGGTATGGTGCTCGCCGGCGTTGGAGTCGGTATCGCTTTGGTTCAGCTCCTCGGTGTGATATTCGCATGCTGCCTCGCTCGTTCCATTCGCAGCCAATATGAAACAGTTTAA
Protein
MFSVFLLIIFVAELAVGIAGYMKHTDLEDSVMRNLNASITQYPVDKNVQKTIDIIQTDLQCCGINSPADWADHGLPIPSTCCSAQEINDGVVAACTENSTNFHSKGCLTKLVVHMKDIGMVLAGVGVGIALVQLLGVIFACCLARSIRSQYETV
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Feature
chain Tetraspanin
Uniprot
Q2F5V7
Q9NB10
Q5MGP8
A0A2H1WWQ3
A0A3S2LWH7
S4PYD2
+ More
A0A194QWI4 A0A194Q3I5 I4DMY1 Q60FR9 A0A1E1W138 I4DPV4 A0A0L7KWS2 A0A212F0C8 E9J782 A0A0B5H7U5 A0A1W4XRX4 A0A2M4C2Z6 A0A182F2L9 W5JDD0 A0A2M4CVN0 A0A2M4BYY8 A0A1B0D6J1 A0A195FSB6 A0A2M4AUK4 A0A2M4CKJ9 A0A2M4BYB1 A0A2M4ATY8 A0A2M3ZCD0 A0A060Q7H8 U5EVW1 A0A195EKQ3 T1EB32 A0A2M4CTM9 A0A1I8M784 A0A2J7QZL2 D6X1Y5 A0A182MUZ1 A0A182VTP8 A0A0T6AV57 A0A182VC99 A0A182Y8G9 A0A182L035 A0A195C3K8 A0A182NB46 A0A182TVX3 A0A182QJI5 Q5TQJ1 A0A182IBU2 A0A088AJM5 A0A2A3E8P8 A0A158NJM3 A0A182JDH2 A0A084WCK9 A0A195B6H4 R4UW62 A0A0L7QP89 A0A023EMW2 A0A1B6H4L4 A0A1L8DK25 A0A1L8DK60 A0A1I8NW60 A0A0J7KBL9 E2A971 A0A1J1ILK4 A0A1B6IF02 A0A210PG06 A0A0P6IVF7 A0A182P4P3 C1C4F6 A0A0B7B6P4 E2BUA7 A0A1Q3G1S2 A0A087TFG6 A0A0A1XA90 A0A0K8TX51 E0VTU3 A0A067QFM7 A0A0C9RNX4 A0A1B6J372 A0A034WFT7 Q9VID1 A0A2C9KTG3 A0A1B6FJP4 A0A1B6MUJ9 T1II96 K7IWT9 A0A1B6KYM3 A0A2I9LNV4 B3NLX2 B3MMB6 B4Q4D8 A0A3Q2XPV4 H7CHJ1 A0A2T7PWD3 T1IN95 A0A1B6KDN1
A0A194QWI4 A0A194Q3I5 I4DMY1 Q60FR9 A0A1E1W138 I4DPV4 A0A0L7KWS2 A0A212F0C8 E9J782 A0A0B5H7U5 A0A1W4XRX4 A0A2M4C2Z6 A0A182F2L9 W5JDD0 A0A2M4CVN0 A0A2M4BYY8 A0A1B0D6J1 A0A195FSB6 A0A2M4AUK4 A0A2M4CKJ9 A0A2M4BYB1 A0A2M4ATY8 A0A2M3ZCD0 A0A060Q7H8 U5EVW1 A0A195EKQ3 T1EB32 A0A2M4CTM9 A0A1I8M784 A0A2J7QZL2 D6X1Y5 A0A182MUZ1 A0A182VTP8 A0A0T6AV57 A0A182VC99 A0A182Y8G9 A0A182L035 A0A195C3K8 A0A182NB46 A0A182TVX3 A0A182QJI5 Q5TQJ1 A0A182IBU2 A0A088AJM5 A0A2A3E8P8 A0A158NJM3 A0A182JDH2 A0A084WCK9 A0A195B6H4 R4UW62 A0A0L7QP89 A0A023EMW2 A0A1B6H4L4 A0A1L8DK25 A0A1L8DK60 A0A1I8NW60 A0A0J7KBL9 E2A971 A0A1J1ILK4 A0A1B6IF02 A0A210PG06 A0A0P6IVF7 A0A182P4P3 C1C4F6 A0A0B7B6P4 E2BUA7 A0A1Q3G1S2 A0A087TFG6 A0A0A1XA90 A0A0K8TX51 E0VTU3 A0A067QFM7 A0A0C9RNX4 A0A1B6J372 A0A034WFT7 Q9VID1 A0A2C9KTG3 A0A1B6FJP4 A0A1B6MUJ9 T1II96 K7IWT9 A0A1B6KYM3 A0A2I9LNV4 B3NLX2 B3MMB6 B4Q4D8 A0A3Q2XPV4 H7CHJ1 A0A2T7PWD3 T1IN95 A0A1B6KDN1
Pubmed
11122467
16023793
23622113
26354079
22651552
26227816
+ More
22118469 21282665 20920257 23761445 25315136 18362917 19820115 25244985 20966253 12364791 14747013 17210077 21347285 24438588 24945155 20798317 28812685 26999592 25830018 20566863 24845553 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15562597 20075255 29248469 17994087 22936249
22118469 21282665 20920257 23761445 25315136 18362917 19820115 25244985 20966253 12364791 14747013 17210077 21347285 24438588 24945155 20798317 28812685 26999592 25830018 20566863 24845553 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15562597 20075255 29248469 17994087 22936249
EMBL
DQ311316
ABD36260.1
AF274021
AAF90147.1
AY829738
AAV91352.1
+ More
ODYU01011572 SOQ57396.1 RSAL01000147 RVE45865.1 GAIX01003493 JAA89067.1 KQ461175 KPJ07931.1 KQ459472 KPJ00103.1 AK402649 BAM19271.1 AB189030 BAD52262.1 GDQN01010346 GDQN01003174 JAT80708.1 JAT87880.1 AK403799 BAM19944.1 JTDY01004985 KOB67504.1 AGBW02011105 OWR47199.1 GL768424 EFZ11326.1 KJ535125 AJF38198.1 GGFJ01010237 MBW59378.1 ADMH02001848 ETN60890.1 GGFL01005216 MBW69394.1 GGFJ01008990 MBW58131.1 AJVK01003639 AJVK01003640 AJVK01003641 AJVK01003642 AJVK01003643 KQ981305 KYN42794.1 GGFK01011133 MBW44454.1 GGFL01001655 MBW65833.1 GGFJ01008908 MBW58049.1 GGFK01010861 MBW44182.1 GGFM01005411 MBW26162.1 HG316497 CDF77374.1 GANO01001692 JAB58179.1 KQ978739 KYN28818.1 GAMD01000403 JAB01188.1 GGFL01004451 MBW68629.1 NEVH01009069 PNF34005.1 KQ971371 EFA10185.1 AXCM01007720 LJIG01022738 KRT78969.1 KQ978292 KYM95449.1 AXCN02000643 AAAB01008964 EAA12176.4 APCN01000738 APCN01000739 APCN01000740 KZ288349 PBC27441.1 ADTU01001946 ADTU01001947 ADTU01001948 ATLV01022709 KE525336 KFB47953.1 KQ976582 KYM79784.1 KC571982 AGM32481.1 KQ414821 KOC60445.1 GAPW01003914 GEHC01000801 JAC09684.1 JAV46844.1 GECZ01000130 JAS69639.1 GFDF01007272 JAV06812.1 GFDF01007258 JAV06826.1 LBMM01009958 KMQ87758.1 GL437711 EFN70056.1 CVRI01000055 CRL01040.1 GECU01022234 JAS85472.1 NEDP02076728 OWF35418.1 GDUN01000926 JAN94993.1 BT081735 ACO51866.1 HACG01040930 CEK87795.1 GL450619 EFN80735.1 GFDL01001278 JAV33767.1 KK114976 KFM63855.1 GBXI01013469 GBXI01006642 GBXI01000751 JAD00823.1 JAD07650.1 JAD13541.1 GDHF01033468 GDHF01008433 GDHF01002375 JAI18846.1 JAI43881.1 JAI49939.1 DS235773 EEB16799.1 KK853603 KDR06366.1 GBYB01012814 GBYB01015237 JAG82581.1 JAG85004.1 GECU01014061 JAS93645.1 GAKP01005394 GAKP01005393 JAC53559.1 AE014134 BT010223 AAF53992.3 AAQ23541.1 GECZ01030572 GECZ01030333 GECZ01025548 GECZ01024288 GECZ01019368 GECZ01016109 GECZ01015276 GECZ01014808 GECZ01009428 GECZ01006774 JAS39197.1 JAS39436.1 JAS44221.1 JAS45481.1 JAS50401.1 JAS53660.1 JAS54493.1 JAS54961.1 JAS60341.1 JAS62995.1 GEBQ01000361 JAT39616.1 JH430149 AAZX01011889 AAZX01017071 GEBQ01023430 JAT16547.1 GFWZ01000074 MBW20064.1 CH954179 EDV54438.1 CH902620 EDV31876.1 CM000361 CM002910 EDX05735.1 KMY91343.1 AB701637 BAL70289.1 PZQS01000001 PVD37729.1 JH431152 GEBQ01030408 JAT09569.1
ODYU01011572 SOQ57396.1 RSAL01000147 RVE45865.1 GAIX01003493 JAA89067.1 KQ461175 KPJ07931.1 KQ459472 KPJ00103.1 AK402649 BAM19271.1 AB189030 BAD52262.1 GDQN01010346 GDQN01003174 JAT80708.1 JAT87880.1 AK403799 BAM19944.1 JTDY01004985 KOB67504.1 AGBW02011105 OWR47199.1 GL768424 EFZ11326.1 KJ535125 AJF38198.1 GGFJ01010237 MBW59378.1 ADMH02001848 ETN60890.1 GGFL01005216 MBW69394.1 GGFJ01008990 MBW58131.1 AJVK01003639 AJVK01003640 AJVK01003641 AJVK01003642 AJVK01003643 KQ981305 KYN42794.1 GGFK01011133 MBW44454.1 GGFL01001655 MBW65833.1 GGFJ01008908 MBW58049.1 GGFK01010861 MBW44182.1 GGFM01005411 MBW26162.1 HG316497 CDF77374.1 GANO01001692 JAB58179.1 KQ978739 KYN28818.1 GAMD01000403 JAB01188.1 GGFL01004451 MBW68629.1 NEVH01009069 PNF34005.1 KQ971371 EFA10185.1 AXCM01007720 LJIG01022738 KRT78969.1 KQ978292 KYM95449.1 AXCN02000643 AAAB01008964 EAA12176.4 APCN01000738 APCN01000739 APCN01000740 KZ288349 PBC27441.1 ADTU01001946 ADTU01001947 ADTU01001948 ATLV01022709 KE525336 KFB47953.1 KQ976582 KYM79784.1 KC571982 AGM32481.1 KQ414821 KOC60445.1 GAPW01003914 GEHC01000801 JAC09684.1 JAV46844.1 GECZ01000130 JAS69639.1 GFDF01007272 JAV06812.1 GFDF01007258 JAV06826.1 LBMM01009958 KMQ87758.1 GL437711 EFN70056.1 CVRI01000055 CRL01040.1 GECU01022234 JAS85472.1 NEDP02076728 OWF35418.1 GDUN01000926 JAN94993.1 BT081735 ACO51866.1 HACG01040930 CEK87795.1 GL450619 EFN80735.1 GFDL01001278 JAV33767.1 KK114976 KFM63855.1 GBXI01013469 GBXI01006642 GBXI01000751 JAD00823.1 JAD07650.1 JAD13541.1 GDHF01033468 GDHF01008433 GDHF01002375 JAI18846.1 JAI43881.1 JAI49939.1 DS235773 EEB16799.1 KK853603 KDR06366.1 GBYB01012814 GBYB01015237 JAG82581.1 JAG85004.1 GECU01014061 JAS93645.1 GAKP01005394 GAKP01005393 JAC53559.1 AE014134 BT010223 AAF53992.3 AAQ23541.1 GECZ01030572 GECZ01030333 GECZ01025548 GECZ01024288 GECZ01019368 GECZ01016109 GECZ01015276 GECZ01014808 GECZ01009428 GECZ01006774 JAS39197.1 JAS39436.1 JAS44221.1 JAS45481.1 JAS50401.1 JAS53660.1 JAS54493.1 JAS54961.1 JAS60341.1 JAS62995.1 GEBQ01000361 JAT39616.1 JH430149 AAZX01011889 AAZX01017071 GEBQ01023430 JAT16547.1 GFWZ01000074 MBW20064.1 CH954179 EDV54438.1 CH902620 EDV31876.1 CM000361 CM002910 EDX05735.1 KMY91343.1 AB701637 BAL70289.1 PZQS01000001 PVD37729.1 JH431152 GEBQ01030408 JAT09569.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000037510
UP000007151
UP000192223
+ More
UP000069272 UP000000673 UP000092462 UP000078541 UP000078492 UP000095301 UP000235965 UP000007266 UP000075883 UP000075920 UP000075903 UP000076408 UP000075882 UP000078542 UP000075884 UP000075902 UP000075886 UP000007062 UP000075840 UP000005203 UP000242457 UP000005205 UP000075880 UP000030765 UP000078540 UP000053825 UP000095300 UP000036403 UP000000311 UP000183832 UP000242188 UP000075885 UP000008237 UP000054359 UP000009046 UP000027135 UP000000803 UP000076420 UP000002358 UP000008711 UP000007801 UP000000304 UP000264820 UP000245119
UP000069272 UP000000673 UP000092462 UP000078541 UP000078492 UP000095301 UP000235965 UP000007266 UP000075883 UP000075920 UP000075903 UP000076408 UP000075882 UP000078542 UP000075884 UP000075902 UP000075886 UP000007062 UP000075840 UP000005203 UP000242457 UP000005205 UP000075880 UP000030765 UP000078540 UP000053825 UP000095300 UP000036403 UP000000311 UP000183832 UP000242188 UP000075885 UP000008237 UP000054359 UP000009046 UP000027135 UP000000803 UP000076420 UP000002358 UP000008711 UP000007801 UP000000304 UP000264820 UP000245119
Pfam
PF00335 Tetraspanin
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
Q2F5V7
Q9NB10
Q5MGP8
A0A2H1WWQ3
A0A3S2LWH7
S4PYD2
+ More
A0A194QWI4 A0A194Q3I5 I4DMY1 Q60FR9 A0A1E1W138 I4DPV4 A0A0L7KWS2 A0A212F0C8 E9J782 A0A0B5H7U5 A0A1W4XRX4 A0A2M4C2Z6 A0A182F2L9 W5JDD0 A0A2M4CVN0 A0A2M4BYY8 A0A1B0D6J1 A0A195FSB6 A0A2M4AUK4 A0A2M4CKJ9 A0A2M4BYB1 A0A2M4ATY8 A0A2M3ZCD0 A0A060Q7H8 U5EVW1 A0A195EKQ3 T1EB32 A0A2M4CTM9 A0A1I8M784 A0A2J7QZL2 D6X1Y5 A0A182MUZ1 A0A182VTP8 A0A0T6AV57 A0A182VC99 A0A182Y8G9 A0A182L035 A0A195C3K8 A0A182NB46 A0A182TVX3 A0A182QJI5 Q5TQJ1 A0A182IBU2 A0A088AJM5 A0A2A3E8P8 A0A158NJM3 A0A182JDH2 A0A084WCK9 A0A195B6H4 R4UW62 A0A0L7QP89 A0A023EMW2 A0A1B6H4L4 A0A1L8DK25 A0A1L8DK60 A0A1I8NW60 A0A0J7KBL9 E2A971 A0A1J1ILK4 A0A1B6IF02 A0A210PG06 A0A0P6IVF7 A0A182P4P3 C1C4F6 A0A0B7B6P4 E2BUA7 A0A1Q3G1S2 A0A087TFG6 A0A0A1XA90 A0A0K8TX51 E0VTU3 A0A067QFM7 A0A0C9RNX4 A0A1B6J372 A0A034WFT7 Q9VID1 A0A2C9KTG3 A0A1B6FJP4 A0A1B6MUJ9 T1II96 K7IWT9 A0A1B6KYM3 A0A2I9LNV4 B3NLX2 B3MMB6 B4Q4D8 A0A3Q2XPV4 H7CHJ1 A0A2T7PWD3 T1IN95 A0A1B6KDN1
A0A194QWI4 A0A194Q3I5 I4DMY1 Q60FR9 A0A1E1W138 I4DPV4 A0A0L7KWS2 A0A212F0C8 E9J782 A0A0B5H7U5 A0A1W4XRX4 A0A2M4C2Z6 A0A182F2L9 W5JDD0 A0A2M4CVN0 A0A2M4BYY8 A0A1B0D6J1 A0A195FSB6 A0A2M4AUK4 A0A2M4CKJ9 A0A2M4BYB1 A0A2M4ATY8 A0A2M3ZCD0 A0A060Q7H8 U5EVW1 A0A195EKQ3 T1EB32 A0A2M4CTM9 A0A1I8M784 A0A2J7QZL2 D6X1Y5 A0A182MUZ1 A0A182VTP8 A0A0T6AV57 A0A182VC99 A0A182Y8G9 A0A182L035 A0A195C3K8 A0A182NB46 A0A182TVX3 A0A182QJI5 Q5TQJ1 A0A182IBU2 A0A088AJM5 A0A2A3E8P8 A0A158NJM3 A0A182JDH2 A0A084WCK9 A0A195B6H4 R4UW62 A0A0L7QP89 A0A023EMW2 A0A1B6H4L4 A0A1L8DK25 A0A1L8DK60 A0A1I8NW60 A0A0J7KBL9 E2A971 A0A1J1ILK4 A0A1B6IF02 A0A210PG06 A0A0P6IVF7 A0A182P4P3 C1C4F6 A0A0B7B6P4 E2BUA7 A0A1Q3G1S2 A0A087TFG6 A0A0A1XA90 A0A0K8TX51 E0VTU3 A0A067QFM7 A0A0C9RNX4 A0A1B6J372 A0A034WFT7 Q9VID1 A0A2C9KTG3 A0A1B6FJP4 A0A1B6MUJ9 T1II96 K7IWT9 A0A1B6KYM3 A0A2I9LNV4 B3NLX2 B3MMB6 B4Q4D8 A0A3Q2XPV4 H7CHJ1 A0A2T7PWD3 T1IN95 A0A1B6KDN1
PDB
2M7Z
E-value=0.0016765,
Score=92
Ontologies
GO
Topology
Subcellular location
Membrane
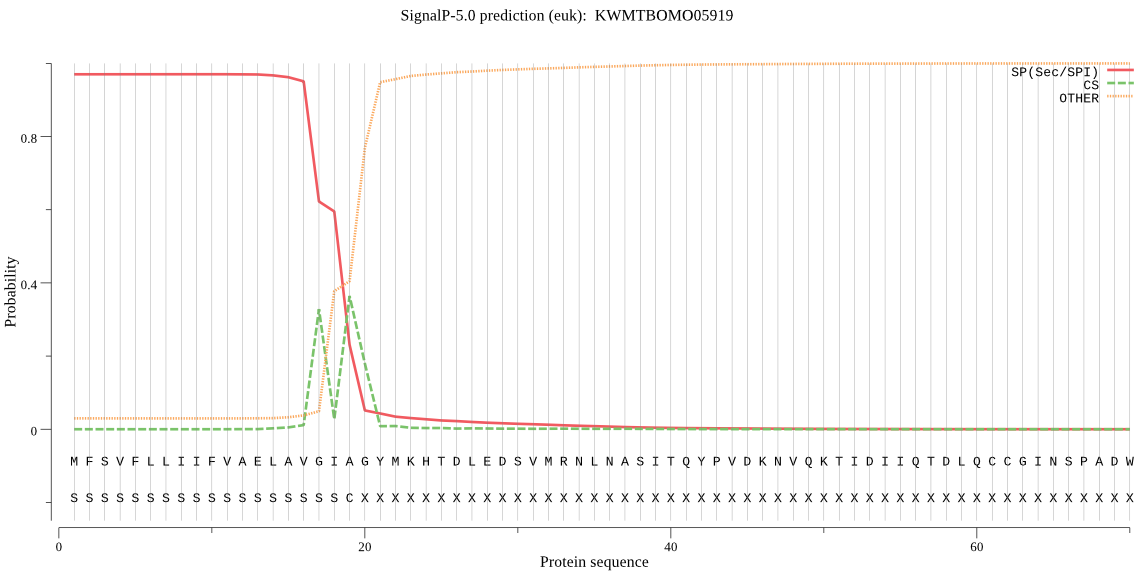
SignalP
Position: 1 - 19,
Likelihood: 0.969984
Length:
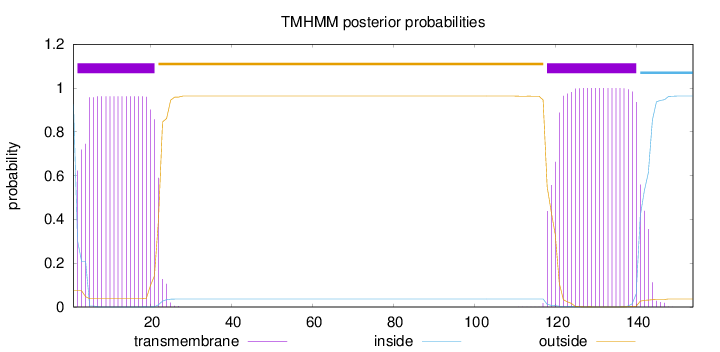
154
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.05316
Exp number, first 60 AAs:
19.1216
Total prob of N-in:
0.92616
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 21
outside
22 - 117
TMhelix
118 - 140
inside
141 - 154
Population Genetic Test Statistics
Pi
226.568633
Theta
187.797017
Tajima's D
0.685364
CLR
0.235595
CSRT
0.567371631418429
Interpretation
Uncertain
