Gene
KWMTBOMO05915
Pre Gene Modal
BGIBMGA002919
Annotation
PREDICTED:_tetraspanin-4-like_[Papilio_polytes]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.901
Sequence
CDS
ATGTTGTCGAAAATATTCACTACGGTGAAATACACATTGGTGACAGTGAATTTTTTGTTCTTGATAACCGGAATTATAATATTGGCTGTCGGAGGCTCGGTGCAAGCTGCCTATAATAATTACCATGAGTTTCTATCGGAGAGGTTTTTCTCTCTTCCCGCCTTTTGTATAGCGACGGGAATTATTATATTCCTCATATCCTTCTCCGGTTTCTATGGAGCCTTCATGGAGAACTACTTAATGAACATGGCGTTTGCGGGTGCCATGATCTTGATGTTCGTTTTTCAACTATCGGCCTGCATAGCTGGTTATGCATTGAAGAGCAACACGGTAGCTTTAGTTCAAAAGCAGCTGTTATCCACCATGGAACTGTACGGACCTGCCTCATTAGAGATCACCAAGCTCTGGGATGAAGTTCAAGGAGACTTCGAATGCTGTGGTGTAGTTAACTCTAGTGACTGGCTTGCGCCCCTCGGGACGCTGGAATCCAGAGGGCTCCCCGTAAGTTGCTGCAGCCACATTTACGGCACCATCCAGCTGTTCAACTGTACGACCGTCGAGGCCTACCCGACTGGTTGTGCCGAAGCGTTCGGTAACTGGGTACAGTCGCACGCGGCGGCAATAGGCGCGGCTGGCATATTCTTAGTTTTAATGCAGGTTTGA
Protein
MLSKIFTTVKYTLVTVNFLFLITGIIILAVGGSVQAAYNNYHEFLSERFFSLPAFCIATGIIIFLISFSGFYGAFMENYLMNMAFAGAMILMFVFQLSACIAGYALKSNTVALVQKQLLSTMELYGPASLEITKLWDEVQGDFECCGVVNSSDWLAPLGTLESRGLPVSCCSHIYGTIQLFNCTTVEAYPTGCAEAFGNWVQSHAAAIGAAGIFLVLMQV
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9J084
A0A2A4IYM5
A0A194QRG1
A0A212F0H8
A0A2H1WHQ5
A0A1E1WEA3
+ More
A0A194Q3J6 A0A3S2LVP5 A0A0L7LID7 A0A336N925 A0A336LVF9 U5EZJ8 B3N7U6 K7IWU0 B4KEY4 A0A3B0JH53 A0A0C9QE23 B0WWM6 A0A1L8DKE1 A0A023EL63 B4N098 A0A1W4VM88 Q1HR03 B4M9P5 B4HYS4 A0A0M4EPZ9 A0A0M5J6B1 B3MM83 Q9VLH1 B4GJ37 H7CHJ1 A0A1A9VA64 A0A0J9QZ16 Q29JV0 B4JP50 A0A1I8NCC7 A0A1I8PKV3 A0A195FQC8 A0A1B0AAK9 D3TNV1 A0A182L665 A0A0L0CFI0 A0A1L8EFT9 B4NXJ3 A0A182RGQ4 A0A182Y8B2 A0A182US04 A0A182ICS9 A0A158NJP6 Q7Q1S3 A0A232FCE8 A0A1A9Y1P1 A0A2M3YZD8 A0A182VTW7 T1DLQ6 A0A182PKV3 A0A1B0D6J2 A0A182JNR5 A0A154PIJ4 A0A084VWC1 A0A2M4BXS0 B6DDW1 W5J645 A0A182F4D2 A0A182NAX7 A0A034WG34 A0A1J1ILC1 A0A182J8J5 A0A2M4A1F7 A0A194QSH1 F4WCE1 A0A1B0B298 A0A182SPQ2 A0A2A3E6W7 V9IFS0 A0A195ELK9 A0A195C5J9 A0A088AJF5 A0A151XK38 A0A1L8E9P8 W8C0N6 A0A1Y1MU82 A0A212F0D6 A0A0K8WI46 B4Q7A8 A0A067QFM7 A0A182WZD8 A0A1Q3FNJ5 A0A1B0CUZ7 A0A0A1WGM2 D6X1Y8 E9J783 A0A182TM64 A0A1E1W325 A0A026WS85 E2A972 A0A182G2C2 A0A182MNY8
A0A194Q3J6 A0A3S2LVP5 A0A0L7LID7 A0A336N925 A0A336LVF9 U5EZJ8 B3N7U6 K7IWU0 B4KEY4 A0A3B0JH53 A0A0C9QE23 B0WWM6 A0A1L8DKE1 A0A023EL63 B4N098 A0A1W4VM88 Q1HR03 B4M9P5 B4HYS4 A0A0M4EPZ9 A0A0M5J6B1 B3MM83 Q9VLH1 B4GJ37 H7CHJ1 A0A1A9VA64 A0A0J9QZ16 Q29JV0 B4JP50 A0A1I8NCC7 A0A1I8PKV3 A0A195FQC8 A0A1B0AAK9 D3TNV1 A0A182L665 A0A0L0CFI0 A0A1L8EFT9 B4NXJ3 A0A182RGQ4 A0A182Y8B2 A0A182US04 A0A182ICS9 A0A158NJP6 Q7Q1S3 A0A232FCE8 A0A1A9Y1P1 A0A2M3YZD8 A0A182VTW7 T1DLQ6 A0A182PKV3 A0A1B0D6J2 A0A182JNR5 A0A154PIJ4 A0A084VWC1 A0A2M4BXS0 B6DDW1 W5J645 A0A182F4D2 A0A182NAX7 A0A034WG34 A0A1J1ILC1 A0A182J8J5 A0A2M4A1F7 A0A194QSH1 F4WCE1 A0A1B0B298 A0A182SPQ2 A0A2A3E6W7 V9IFS0 A0A195ELK9 A0A195C5J9 A0A088AJF5 A0A151XK38 A0A1L8E9P8 W8C0N6 A0A1Y1MU82 A0A212F0D6 A0A0K8WI46 B4Q7A8 A0A067QFM7 A0A182WZD8 A0A1Q3FNJ5 A0A1B0CUZ7 A0A0A1WGM2 D6X1Y8 E9J783 A0A182TM64 A0A1E1W325 A0A026WS85 E2A972 A0A182G2C2 A0A182MNY8
Pubmed
19121390
26354079
22118469
26227816
17994087
18057021
+ More
20075255 24945155 17204158 11122467 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 25315136 20353571 20966253 26108605 17550304 25244985 21347285 12364791 14747013 17210077 28648823 24438588 19178717 20920257 23761445 25348373 21719571 24495485 28004739 24845553 25830018 18362917 19820115 21282665 24508170 20798317 26483478
20075255 24945155 17204158 11122467 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 25315136 20353571 20966253 26108605 17550304 25244985 21347285 12364791 14747013 17210077 28648823 24438588 19178717 20920257 23761445 25348373 21719571 24495485 28004739 24845553 25830018 18362917 19820115 21282665 24508170 20798317 26483478
EMBL
BABH01019084
BABH01019085
NWSH01004989
PCG64518.1
KQ461175
KPJ07929.1
+ More
AGBW02011105 OWR47201.1 ODYU01008695 SOQ52486.1 GDQN01005730 GDQN01001844 JAT85324.1 JAT89210.1 KQ459472 KPJ00102.1 RSAL01000003 RVE54769.1 JTDY01000950 KOB75323.1 UFQS01003761 UFQT01003761 SSX15916.1 SSX35258.1 UFQT01000215 SSX21810.1 GANO01000667 JAB59204.1 CH954177 EDV58307.1 AAZX01000576 CH933807 EDW11953.1 KRG02996.1 OUUW01000006 SPP81515.1 GBYB01012658 JAG82425.1 DS232149 EDS36118.1 GFDF01007247 JAV06837.1 GAPW01003665 GAPW01003664 GAPW01003663 GAPW01003662 JAC09935.1 CH963920 EDW77511.1 KRF98688.1 DQ440291 ABF18324.1 CH940654 EDW57921.1 KRF78024.1 CH480818 EDW52204.1 CP012523 ALC39012.1 ALC39536.1 CH902620 EDV31843.1 KPU73654.1 AE014134 AF274011 AY071356 AAF52719.2 AAF90137.1 AAL48978.1 ACZ94205.1 CH479184 EDW37351.1 AB701637 BAL70289.1 CM002910 KMY89176.1 CH379062 EAL32861.1 KRT05022.1 CH916372 EDV99475.1 KQ981305 KYN42795.1 EZ423103 ADD19379.1 JRES01000477 KNC30957.1 GFDG01001330 JAV17469.1 CM000157 EDW88584.1 APCN01005756 ADTU01001950 AAAB01008980 EAA13868.2 NNAY01000426 OXU28496.1 GGFM01000872 MBW21623.1 GAMD01000291 JAB01300.1 AJVK01003646 AJVK01003647 AJVK01003648 AJVK01003649 KQ434924 KZC11627.1 ATLV01017552 KE525174 KFB42265.1 GGFJ01008709 MBW57850.1 EU934330 ACI30108.1 ADMH02002138 ETN58335.1 GAKP01005680 GAKP01005679 JAC53272.1 CVRI01000055 CRL01039.1 GGFK01001302 MBW34623.1 KPJ07930.1 GL888070 EGI68121.1 JXJN01007522 JXJN01007523 KZ288349 PBC27440.1 JR040956 AEY59336.1 KQ978739 KYN28817.1 KQ978292 KYM95448.1 KQ982052 KYQ60665.1 GFDG01003342 JAV15457.1 GAMC01003917 GAMC01003916 JAC02639.1 GEZM01025139 GEZM01025138 JAV87566.1 OWR47200.1 GDHF01024480 GDHF01011543 GDHF01001754 JAI27834.1 JAI40771.1 JAI50560.1 CM000361 EDX04302.1 KK853603 KDR06366.1 GFDL01005875 JAV29170.1 AJWK01029905 AJWK01029906 GBXI01016642 JAC97649.1 KQ971371 EFA10187.1 GL768424 EFZ11325.1 GDQN01009677 JAT81377.1 KK107119 EZA58823.1 GL437711 EFN70057.1 JXUM01139034 JXUM01139035 JXUM01139036 JXUM01139037 AXCM01001829
AGBW02011105 OWR47201.1 ODYU01008695 SOQ52486.1 GDQN01005730 GDQN01001844 JAT85324.1 JAT89210.1 KQ459472 KPJ00102.1 RSAL01000003 RVE54769.1 JTDY01000950 KOB75323.1 UFQS01003761 UFQT01003761 SSX15916.1 SSX35258.1 UFQT01000215 SSX21810.1 GANO01000667 JAB59204.1 CH954177 EDV58307.1 AAZX01000576 CH933807 EDW11953.1 KRG02996.1 OUUW01000006 SPP81515.1 GBYB01012658 JAG82425.1 DS232149 EDS36118.1 GFDF01007247 JAV06837.1 GAPW01003665 GAPW01003664 GAPW01003663 GAPW01003662 JAC09935.1 CH963920 EDW77511.1 KRF98688.1 DQ440291 ABF18324.1 CH940654 EDW57921.1 KRF78024.1 CH480818 EDW52204.1 CP012523 ALC39012.1 ALC39536.1 CH902620 EDV31843.1 KPU73654.1 AE014134 AF274011 AY071356 AAF52719.2 AAF90137.1 AAL48978.1 ACZ94205.1 CH479184 EDW37351.1 AB701637 BAL70289.1 CM002910 KMY89176.1 CH379062 EAL32861.1 KRT05022.1 CH916372 EDV99475.1 KQ981305 KYN42795.1 EZ423103 ADD19379.1 JRES01000477 KNC30957.1 GFDG01001330 JAV17469.1 CM000157 EDW88584.1 APCN01005756 ADTU01001950 AAAB01008980 EAA13868.2 NNAY01000426 OXU28496.1 GGFM01000872 MBW21623.1 GAMD01000291 JAB01300.1 AJVK01003646 AJVK01003647 AJVK01003648 AJVK01003649 KQ434924 KZC11627.1 ATLV01017552 KE525174 KFB42265.1 GGFJ01008709 MBW57850.1 EU934330 ACI30108.1 ADMH02002138 ETN58335.1 GAKP01005680 GAKP01005679 JAC53272.1 CVRI01000055 CRL01039.1 GGFK01001302 MBW34623.1 KPJ07930.1 GL888070 EGI68121.1 JXJN01007522 JXJN01007523 KZ288349 PBC27440.1 JR040956 AEY59336.1 KQ978739 KYN28817.1 KQ978292 KYM95448.1 KQ982052 KYQ60665.1 GFDG01003342 JAV15457.1 GAMC01003917 GAMC01003916 JAC02639.1 GEZM01025139 GEZM01025138 JAV87566.1 OWR47200.1 GDHF01024480 GDHF01011543 GDHF01001754 JAI27834.1 JAI40771.1 JAI50560.1 CM000361 EDX04302.1 KK853603 KDR06366.1 GFDL01005875 JAV29170.1 AJWK01029905 AJWK01029906 GBXI01016642 JAC97649.1 KQ971371 EFA10187.1 GL768424 EFZ11325.1 GDQN01009677 JAT81377.1 KK107119 EZA58823.1 GL437711 EFN70057.1 JXUM01139034 JXUM01139035 JXUM01139036 JXUM01139037 AXCM01001829
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000283053
+ More
UP000037510 UP000008711 UP000002358 UP000009192 UP000268350 UP000002320 UP000007798 UP000192221 UP000008792 UP000001292 UP000092553 UP000007801 UP000000803 UP000008744 UP000078200 UP000001819 UP000001070 UP000095301 UP000095300 UP000078541 UP000092445 UP000075882 UP000037069 UP000002282 UP000075900 UP000076408 UP000075903 UP000075840 UP000005205 UP000007062 UP000215335 UP000092443 UP000075920 UP000075885 UP000092462 UP000075881 UP000076502 UP000030765 UP000000673 UP000069272 UP000075884 UP000183832 UP000075880 UP000007755 UP000092460 UP000075901 UP000242457 UP000078492 UP000078542 UP000005203 UP000075809 UP000000304 UP000027135 UP000076407 UP000092461 UP000007266 UP000075902 UP000053097 UP000000311 UP000069940 UP000075883
UP000037510 UP000008711 UP000002358 UP000009192 UP000268350 UP000002320 UP000007798 UP000192221 UP000008792 UP000001292 UP000092553 UP000007801 UP000000803 UP000008744 UP000078200 UP000001819 UP000001070 UP000095301 UP000095300 UP000078541 UP000092445 UP000075882 UP000037069 UP000002282 UP000075900 UP000076408 UP000075903 UP000075840 UP000005205 UP000007062 UP000215335 UP000092443 UP000075920 UP000075885 UP000092462 UP000075881 UP000076502 UP000030765 UP000000673 UP000069272 UP000075884 UP000183832 UP000075880 UP000007755 UP000092460 UP000075901 UP000242457 UP000078492 UP000078542 UP000005203 UP000075809 UP000000304 UP000027135 UP000076407 UP000092461 UP000007266 UP000075902 UP000053097 UP000000311 UP000069940 UP000075883
PRIDE
Interpro
IPR008952
Tetraspanin_EC2_sf
+ More
IPR018499 Tetraspanin/Peripherin
IPR000301 Tetraspanin
IPR018503 Tetraspanin_CS
IPR009049 Argininosuccinate_lyase
IPR022761 Fumarate_lyase_N
IPR020557 Fumarate_lyase_CS
IPR000362 Fumarate_lyase_fam
IPR024083 Fumarase/histidase_N
IPR029419 Arg_succ_lyase_C
IPR008948 L-Aspartase-like
IPR018499 Tetraspanin/Peripherin
IPR000301 Tetraspanin
IPR018503 Tetraspanin_CS
IPR009049 Argininosuccinate_lyase
IPR022761 Fumarate_lyase_N
IPR020557 Fumarate_lyase_CS
IPR000362 Fumarate_lyase_fam
IPR024083 Fumarase/histidase_N
IPR029419 Arg_succ_lyase_C
IPR008948 L-Aspartase-like
Gene 3D
ProteinModelPortal
H9J084
A0A2A4IYM5
A0A194QRG1
A0A212F0H8
A0A2H1WHQ5
A0A1E1WEA3
+ More
A0A194Q3J6 A0A3S2LVP5 A0A0L7LID7 A0A336N925 A0A336LVF9 U5EZJ8 B3N7U6 K7IWU0 B4KEY4 A0A3B0JH53 A0A0C9QE23 B0WWM6 A0A1L8DKE1 A0A023EL63 B4N098 A0A1W4VM88 Q1HR03 B4M9P5 B4HYS4 A0A0M4EPZ9 A0A0M5J6B1 B3MM83 Q9VLH1 B4GJ37 H7CHJ1 A0A1A9VA64 A0A0J9QZ16 Q29JV0 B4JP50 A0A1I8NCC7 A0A1I8PKV3 A0A195FQC8 A0A1B0AAK9 D3TNV1 A0A182L665 A0A0L0CFI0 A0A1L8EFT9 B4NXJ3 A0A182RGQ4 A0A182Y8B2 A0A182US04 A0A182ICS9 A0A158NJP6 Q7Q1S3 A0A232FCE8 A0A1A9Y1P1 A0A2M3YZD8 A0A182VTW7 T1DLQ6 A0A182PKV3 A0A1B0D6J2 A0A182JNR5 A0A154PIJ4 A0A084VWC1 A0A2M4BXS0 B6DDW1 W5J645 A0A182F4D2 A0A182NAX7 A0A034WG34 A0A1J1ILC1 A0A182J8J5 A0A2M4A1F7 A0A194QSH1 F4WCE1 A0A1B0B298 A0A182SPQ2 A0A2A3E6W7 V9IFS0 A0A195ELK9 A0A195C5J9 A0A088AJF5 A0A151XK38 A0A1L8E9P8 W8C0N6 A0A1Y1MU82 A0A212F0D6 A0A0K8WI46 B4Q7A8 A0A067QFM7 A0A182WZD8 A0A1Q3FNJ5 A0A1B0CUZ7 A0A0A1WGM2 D6X1Y8 E9J783 A0A182TM64 A0A1E1W325 A0A026WS85 E2A972 A0A182G2C2 A0A182MNY8
A0A194Q3J6 A0A3S2LVP5 A0A0L7LID7 A0A336N925 A0A336LVF9 U5EZJ8 B3N7U6 K7IWU0 B4KEY4 A0A3B0JH53 A0A0C9QE23 B0WWM6 A0A1L8DKE1 A0A023EL63 B4N098 A0A1W4VM88 Q1HR03 B4M9P5 B4HYS4 A0A0M4EPZ9 A0A0M5J6B1 B3MM83 Q9VLH1 B4GJ37 H7CHJ1 A0A1A9VA64 A0A0J9QZ16 Q29JV0 B4JP50 A0A1I8NCC7 A0A1I8PKV3 A0A195FQC8 A0A1B0AAK9 D3TNV1 A0A182L665 A0A0L0CFI0 A0A1L8EFT9 B4NXJ3 A0A182RGQ4 A0A182Y8B2 A0A182US04 A0A182ICS9 A0A158NJP6 Q7Q1S3 A0A232FCE8 A0A1A9Y1P1 A0A2M3YZD8 A0A182VTW7 T1DLQ6 A0A182PKV3 A0A1B0D6J2 A0A182JNR5 A0A154PIJ4 A0A084VWC1 A0A2M4BXS0 B6DDW1 W5J645 A0A182F4D2 A0A182NAX7 A0A034WG34 A0A1J1ILC1 A0A182J8J5 A0A2M4A1F7 A0A194QSH1 F4WCE1 A0A1B0B298 A0A182SPQ2 A0A2A3E6W7 V9IFS0 A0A195ELK9 A0A195C5J9 A0A088AJF5 A0A151XK38 A0A1L8E9P8 W8C0N6 A0A1Y1MU82 A0A212F0D6 A0A0K8WI46 B4Q7A8 A0A067QFM7 A0A182WZD8 A0A1Q3FNJ5 A0A1B0CUZ7 A0A0A1WGM2 D6X1Y8 E9J783 A0A182TM64 A0A1E1W325 A0A026WS85 E2A972 A0A182G2C2 A0A182MNY8
Ontologies
GO
PANTHER
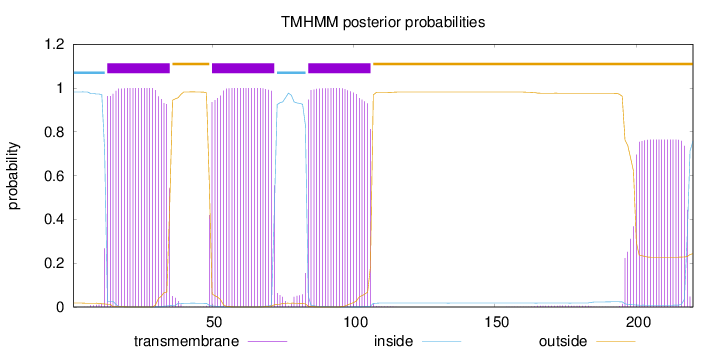
Topology
Subcellular location
Membrane
Length:
220
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
83.82183
Exp number, first 60 AAs:
33.84473
Total prob of N-in:
0.98230
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 49
TMhelix
50 - 72
inside
73 - 83
TMhelix
84 - 106
outside
107 - 220
Population Genetic Test Statistics
Pi
24.79666
Theta
186.932864
Tajima's D
1.12211
CLR
0.053677
CSRT
0.699615019249038
Interpretation
Uncertain
