Gene
KWMTBOMO05914 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002920
Annotation
PREDICTED:_CD63_antigen-like_[Papilio_polytes]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 3.636
Sequence
CDS
ATGCTGTTCGTCTCTCTCTTCGGCTGCATCGGTGCCCTGAAAGAGAGTACCTGCTTGGTCAACATTTTCGCAGTGATCCTTAGCATCGTGTTCATTCTGGAGATCGCGGCGGCCATCACTGCCTACAGTCTTCGCAGCGAGGTCTCACAATTTCTGGACAAAAAACTTAGGGAAGCACTTCCTCTATACTACCAGCATGCTGAGGCGCAGGATGCATTTGACTTCATTCAATACGAGTTGAACTGCTGCGGCGTCGATAGCTACATGGACTGGCATGCGGTCGACCCCCCCACTGGAGGGACCGGCATTAGCTTTGCCAACCTCACGGTCCCCGATTCGTGCTGCGCCCAGTCCCACATGACTGTCGTCGGTGAAGAGACGATTGTAGAGTGCCATAAGATCTATGCCAACGGTTGCCTGCCCAGAGTCTATTATTTGGTTAACCAAAGCGCTGGACTACTTGGAGCTGGCGCGTTGACCATCTGCTTCATTCAGATAATCGGAATCGTTTTCTCGTTCTCACTGGCGAGCTCTATCCGTAAAGCCAAGATCGAACGCGAACGCAGGCGCTGGGAGATCCAGGAGCGGATGATCAACGCCCACACTTCCTTGAACCCTAACAACGAGAAAGTCGCCCCGGTGGTGTACGTTCCGTTCCAAGGACAGGCCGGCGCTTAA
Protein
MLFVSLFGCIGALKESTCLVNIFAVILSIVFILEIAAAITAYSLRSEVSQFLDKKLREALPLYYQHAEAQDAFDFIQYELNCCGVDSYMDWHAVDPPTGGTGISFANLTVPDSCCAQSHMTVVGEETIVECHKIYANGCLPRVYYLVNQSAGLLGAGALTICFIQIIGIVFSFSLASSIRKAKIERERRRWEIQERMINAHTSLNPNNEKVAPVVYVPFQGQAGA
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9J085
A0A2A4J4A5
A0A2A4IVI7
A0A194QQZ4
A0A194Q9L4
A0A212F0E7
+ More
S4NX85 Q9NB09 A0A3S2PLD7 A0A2H1WHE8 A0A2A4IU03 A0A3S2KXX3 Q16LT3 A0A023EMK4 A0A182H2Z1 A0A023EPW8 A0A023ENQ5 A0A023EMJ0 A0A023ESE8 B0W1E6 A0A1Q3FKK5 A0A1B6KDN1 A0A1Y1K6J6 A0A1B6MCQ8 A0A1J1HRI6 A0A1L8DK41 A0A1B6HCW8 A0A1L8DJV1 A0A1L8DJK5 A0A2J7QZJ0 A0A0J7L6K8 A0A1L8DKC0 A0A1B6HHR4 A0A1Y1K6D0 A0A1B6GIS6 D6X0E2 A0A1B6FPI6 W8AVJ1 A0A0A1X025 A0A1B0D6J3 U5EP47 A0A151XJT2 E2A973 A0A1B0CUZ7 A0A182IPX2 A0A067QFM7 A0A182RK40 W8ARL8 A0A3M0IVD9 A0A0K8W976 A0A0K8VE31 J3JUM3 A0A1S4GZL2 A0A182X5V5 A0A182IBI8 A0A182LRX5 C3UTC3 A0A182SHP8 A0A182NGQ4 A0A182WFF0 A0A182K5V7 A0A034W8M1 A0A0L0BXQ5 B4Q7A9 A0A195C3M0 A0A182P1L4 E9J782 A0A1A9W2K0 C1C4F6 A0A182YN77 A0A182QIV2 B4NXJ4 A0A3B3RV58 A0A1B0B299 A0A3B1IPS9 A0A3B3RV68 A0A158NJN4 A0A1L8EF34 A0A1L8EAQ7 B4KEY5 A0A1A9Y1P2 Q9VLH0 K1QBC6 A0A232FCT9 A0A1B0AAL1 A0A1A9VA65 D3TQ22 B4HYS5 A0A1V4J584 B3N7U7 F4WCE0 A0A195B5J4 A0A1W4VMH2 T1P8S2 A0A3B1J8E6 A0A0K8TPC7 A0A3N0YM71 A0A067QFC7 A0A1B6E2I7
S4NX85 Q9NB09 A0A3S2PLD7 A0A2H1WHE8 A0A2A4IU03 A0A3S2KXX3 Q16LT3 A0A023EMK4 A0A182H2Z1 A0A023EPW8 A0A023ENQ5 A0A023EMJ0 A0A023ESE8 B0W1E6 A0A1Q3FKK5 A0A1B6KDN1 A0A1Y1K6J6 A0A1B6MCQ8 A0A1J1HRI6 A0A1L8DK41 A0A1B6HCW8 A0A1L8DJV1 A0A1L8DJK5 A0A2J7QZJ0 A0A0J7L6K8 A0A1L8DKC0 A0A1B6HHR4 A0A1Y1K6D0 A0A1B6GIS6 D6X0E2 A0A1B6FPI6 W8AVJ1 A0A0A1X025 A0A1B0D6J3 U5EP47 A0A151XJT2 E2A973 A0A1B0CUZ7 A0A182IPX2 A0A067QFM7 A0A182RK40 W8ARL8 A0A3M0IVD9 A0A0K8W976 A0A0K8VE31 J3JUM3 A0A1S4GZL2 A0A182X5V5 A0A182IBI8 A0A182LRX5 C3UTC3 A0A182SHP8 A0A182NGQ4 A0A182WFF0 A0A182K5V7 A0A034W8M1 A0A0L0BXQ5 B4Q7A9 A0A195C3M0 A0A182P1L4 E9J782 A0A1A9W2K0 C1C4F6 A0A182YN77 A0A182QIV2 B4NXJ4 A0A3B3RV58 A0A1B0B299 A0A3B1IPS9 A0A3B3RV68 A0A158NJN4 A0A1L8EF34 A0A1L8EAQ7 B4KEY5 A0A1A9Y1P2 Q9VLH0 K1QBC6 A0A232FCT9 A0A1B0AAL1 A0A1A9VA65 D3TQ22 B4HYS5 A0A1V4J584 B3N7U7 F4WCE0 A0A195B5J4 A0A1W4VMH2 T1P8S2 A0A3B1J8E6 A0A0K8TPC7 A0A3N0YM71 A0A067QFC7 A0A1B6E2I7
Pubmed
19121390
26354079
22118469
23622113
11122467
17510324
+ More
24945155 26483478 28004739 18362917 19820115 24495485 25830018 20798317 24845553 22516182 23537049 12364791 19364528 25348373 26108605 17994087 22936249 21282665 25244985 17550304 29240929 25329095 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22992520 28648823 20353571 21719571 25315136 26369729
24945155 26483478 28004739 18362917 19820115 24495485 25830018 20798317 24845553 22516182 23537049 12364791 19364528 25348373 26108605 17994087 22936249 21282665 25244985 17550304 29240929 25329095 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22992520 28648823 20353571 21719571 25315136 26369729
EMBL
BABH01019090
NWSH01003126
PCG66905.1
NWSH01007000
PCG63160.1
KQ461175
+ More
KPJ07928.1 KQ459472 KPJ00101.1 AGBW02011105 OWR47202.1 GAIX01012262 JAA80298.1 AF274022 AAF90148.1 RSAL01000003 RVE54770.1 ODYU01008695 SOQ52488.1 PCG63159.1 RSAL01000945 RVE41032.1 CH477891 CH477347 EAT35294.1 EAT42969.1 GAPW01003030 JAC10568.1 JXUM01106293 JXUM01106294 KQ565042 KXJ71418.1 GAPW01003029 JAC10569.1 GAPW01003028 JAC10570.1 GAPW01003045 JAC10553.1 GAPW01001917 JAC11681.1 DS231821 EDS45016.1 GFDL01007003 JAV28042.1 GEBQ01030408 JAT09569.1 GEZM01091225 GEZM01091224 GEZM01091223 GEZM01091222 JAV57013.1 GEBQ01006270 JAT33707.1 CVRI01000010 CRK88809.1 GFDF01007246 JAV06838.1 GECU01035122 GECU01005586 JAS72584.1 JAT02121.1 GFDF01007437 JAV06647.1 GFDF01007436 JAV06648.1 NEVH01009069 PNF34006.1 LBMM01000455 KMQ98296.1 GFDF01007259 JAV06825.1 GECU01033473 JAS74233.1 JAV57012.1 GECZ01007431 JAS62338.1 KQ971372 EFA09586.1 GECZ01017644 JAS52125.1 GAMC01017840 GAMC01017837 JAB88718.1 GBXI01011327 GBXI01009628 JAD02965.1 JAD04664.1 AJVK01003649 GANO01000316 JAB59555.1 KQ982052 KYQ60664.1 GL437711 EFN70058.1 AJWK01029905 AJWK01029906 KK853603 KDR06366.1 GAMC01017838 JAB88717.1 QRBI01000226 RMB92422.1 GDHF01006425 GDHF01004904 JAI45889.1 JAI47410.1 GDHF01015183 JAI37131.1 APGK01003084 APGK01003085 APGK01018485 APGK01018486 APGK01018487 BT126938 KB740076 KB735111 KB631643 AEE61900.1 ENN81658.1 ENN83444.1 ERL84920.1 AAAB01008944 APCN01000703 AXCM01001804 FJ654711 ACP18832.1 GAKP01007898 GAKP01007897 JAC51054.1 JRES01001270 KNC24049.1 CM000361 CM002910 EDX04303.1 KMY89177.1 KQ978292 KYM95447.1 GL768424 EFZ11326.1 BT081735 ACO51866.1 AXCN02002079 CM000157 EDW88585.1 JXJN01007521 ADTU01001951 GFDG01001477 JAV17322.1 GFDG01002972 JAV15827.1 CH933807 EDW11954.1 AE014134 AF274012 AY122216 AAF52720.1 AAF90138.1 AAM52728.1 JH818108 EKC18846.1 NNAY01000426 OXU28495.1 CCAG010023894 EZ423524 ADD19800.1 CH480818 EDW52205.1 LSYS01009212 OPJ67316.1 CH954177 EDV58308.1 GL888070 EGI68120.1 KQ976582 KYM79783.1 KA644525 AFP59154.1 GDAI01001361 JAI16242.1 RJVU01036174 ROL47010.1 KDR06365.1 GEDC01031511 GEDC01024465 GEDC01019324 GEDC01018344 GEDC01005153 GEDC01000862 JAS05787.1 JAS12833.1 JAS17974.1 JAS18954.1 JAS32145.1 JAS36436.1
KPJ07928.1 KQ459472 KPJ00101.1 AGBW02011105 OWR47202.1 GAIX01012262 JAA80298.1 AF274022 AAF90148.1 RSAL01000003 RVE54770.1 ODYU01008695 SOQ52488.1 PCG63159.1 RSAL01000945 RVE41032.1 CH477891 CH477347 EAT35294.1 EAT42969.1 GAPW01003030 JAC10568.1 JXUM01106293 JXUM01106294 KQ565042 KXJ71418.1 GAPW01003029 JAC10569.1 GAPW01003028 JAC10570.1 GAPW01003045 JAC10553.1 GAPW01001917 JAC11681.1 DS231821 EDS45016.1 GFDL01007003 JAV28042.1 GEBQ01030408 JAT09569.1 GEZM01091225 GEZM01091224 GEZM01091223 GEZM01091222 JAV57013.1 GEBQ01006270 JAT33707.1 CVRI01000010 CRK88809.1 GFDF01007246 JAV06838.1 GECU01035122 GECU01005586 JAS72584.1 JAT02121.1 GFDF01007437 JAV06647.1 GFDF01007436 JAV06648.1 NEVH01009069 PNF34006.1 LBMM01000455 KMQ98296.1 GFDF01007259 JAV06825.1 GECU01033473 JAS74233.1 JAV57012.1 GECZ01007431 JAS62338.1 KQ971372 EFA09586.1 GECZ01017644 JAS52125.1 GAMC01017840 GAMC01017837 JAB88718.1 GBXI01011327 GBXI01009628 JAD02965.1 JAD04664.1 AJVK01003649 GANO01000316 JAB59555.1 KQ982052 KYQ60664.1 GL437711 EFN70058.1 AJWK01029905 AJWK01029906 KK853603 KDR06366.1 GAMC01017838 JAB88717.1 QRBI01000226 RMB92422.1 GDHF01006425 GDHF01004904 JAI45889.1 JAI47410.1 GDHF01015183 JAI37131.1 APGK01003084 APGK01003085 APGK01018485 APGK01018486 APGK01018487 BT126938 KB740076 KB735111 KB631643 AEE61900.1 ENN81658.1 ENN83444.1 ERL84920.1 AAAB01008944 APCN01000703 AXCM01001804 FJ654711 ACP18832.1 GAKP01007898 GAKP01007897 JAC51054.1 JRES01001270 KNC24049.1 CM000361 CM002910 EDX04303.1 KMY89177.1 KQ978292 KYM95447.1 GL768424 EFZ11326.1 BT081735 ACO51866.1 AXCN02002079 CM000157 EDW88585.1 JXJN01007521 ADTU01001951 GFDG01001477 JAV17322.1 GFDG01002972 JAV15827.1 CH933807 EDW11954.1 AE014134 AF274012 AY122216 AAF52720.1 AAF90138.1 AAM52728.1 JH818108 EKC18846.1 NNAY01000426 OXU28495.1 CCAG010023894 EZ423524 ADD19800.1 CH480818 EDW52205.1 LSYS01009212 OPJ67316.1 CH954177 EDV58308.1 GL888070 EGI68120.1 KQ976582 KYM79783.1 KA644525 AFP59154.1 GDAI01001361 JAI16242.1 RJVU01036174 ROL47010.1 KDR06365.1 GEDC01031511 GEDC01024465 GEDC01019324 GEDC01018344 GEDC01005153 GEDC01000862 JAS05787.1 JAS12833.1 JAS17974.1 JAS18954.1 JAS32145.1 JAS36436.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000183832 UP000235965 UP000036403 UP000007266 UP000092462 UP000075809 UP000000311 UP000092461 UP000075880 UP000027135 UP000075900 UP000269221 UP000019118 UP000030742 UP000076407 UP000075840 UP000075883 UP000075901 UP000075884 UP000075920 UP000075881 UP000037069 UP000000304 UP000078542 UP000075885 UP000091820 UP000076408 UP000075886 UP000002282 UP000261540 UP000092460 UP000018467 UP000005205 UP000009192 UP000092443 UP000000803 UP000005408 UP000215335 UP000092445 UP000078200 UP000092444 UP000001292 UP000190648 UP000008711 UP000007755 UP000078540 UP000192221 UP000095301
UP000008820 UP000069940 UP000249989 UP000002320 UP000183832 UP000235965 UP000036403 UP000007266 UP000092462 UP000075809 UP000000311 UP000092461 UP000075880 UP000027135 UP000075900 UP000269221 UP000019118 UP000030742 UP000076407 UP000075840 UP000075883 UP000075901 UP000075884 UP000075920 UP000075881 UP000037069 UP000000304 UP000078542 UP000075885 UP000091820 UP000076408 UP000075886 UP000002282 UP000261540 UP000092460 UP000018467 UP000005205 UP000009192 UP000092443 UP000000803 UP000005408 UP000215335 UP000092445 UP000078200 UP000092444 UP000001292 UP000190648 UP000008711 UP000007755 UP000078540 UP000192221 UP000095301
Interpro
IPR000301
Tetraspanin
+ More
IPR018499 Tetraspanin/Peripherin
IPR018503 Tetraspanin_CS
IPR008952 Tetraspanin_EC2_sf
IPR007889 HTH_Psq
IPR009057 Homeobox-like_sf
IPR000048 IQ_motif_EF-hand-BS
IPR009049 Argininosuccinate_lyase
IPR022761 Fumarate_lyase_N
IPR020557 Fumarate_lyase_CS
IPR000362 Fumarate_lyase_fam
IPR024083 Fumarase/histidase_N
IPR029419 Arg_succ_lyase_C
IPR008948 L-Aspartase-like
IPR018499 Tetraspanin/Peripherin
IPR018503 Tetraspanin_CS
IPR008952 Tetraspanin_EC2_sf
IPR007889 HTH_Psq
IPR009057 Homeobox-like_sf
IPR000048 IQ_motif_EF-hand-BS
IPR009049 Argininosuccinate_lyase
IPR022761 Fumarate_lyase_N
IPR020557 Fumarate_lyase_CS
IPR000362 Fumarate_lyase_fam
IPR024083 Fumarase/histidase_N
IPR029419 Arg_succ_lyase_C
IPR008948 L-Aspartase-like
Gene 3D
ProteinModelPortal
H9J085
A0A2A4J4A5
A0A2A4IVI7
A0A194QQZ4
A0A194Q9L4
A0A212F0E7
+ More
S4NX85 Q9NB09 A0A3S2PLD7 A0A2H1WHE8 A0A2A4IU03 A0A3S2KXX3 Q16LT3 A0A023EMK4 A0A182H2Z1 A0A023EPW8 A0A023ENQ5 A0A023EMJ0 A0A023ESE8 B0W1E6 A0A1Q3FKK5 A0A1B6KDN1 A0A1Y1K6J6 A0A1B6MCQ8 A0A1J1HRI6 A0A1L8DK41 A0A1B6HCW8 A0A1L8DJV1 A0A1L8DJK5 A0A2J7QZJ0 A0A0J7L6K8 A0A1L8DKC0 A0A1B6HHR4 A0A1Y1K6D0 A0A1B6GIS6 D6X0E2 A0A1B6FPI6 W8AVJ1 A0A0A1X025 A0A1B0D6J3 U5EP47 A0A151XJT2 E2A973 A0A1B0CUZ7 A0A182IPX2 A0A067QFM7 A0A182RK40 W8ARL8 A0A3M0IVD9 A0A0K8W976 A0A0K8VE31 J3JUM3 A0A1S4GZL2 A0A182X5V5 A0A182IBI8 A0A182LRX5 C3UTC3 A0A182SHP8 A0A182NGQ4 A0A182WFF0 A0A182K5V7 A0A034W8M1 A0A0L0BXQ5 B4Q7A9 A0A195C3M0 A0A182P1L4 E9J782 A0A1A9W2K0 C1C4F6 A0A182YN77 A0A182QIV2 B4NXJ4 A0A3B3RV58 A0A1B0B299 A0A3B1IPS9 A0A3B3RV68 A0A158NJN4 A0A1L8EF34 A0A1L8EAQ7 B4KEY5 A0A1A9Y1P2 Q9VLH0 K1QBC6 A0A232FCT9 A0A1B0AAL1 A0A1A9VA65 D3TQ22 B4HYS5 A0A1V4J584 B3N7U7 F4WCE0 A0A195B5J4 A0A1W4VMH2 T1P8S2 A0A3B1J8E6 A0A0K8TPC7 A0A3N0YM71 A0A067QFC7 A0A1B6E2I7
S4NX85 Q9NB09 A0A3S2PLD7 A0A2H1WHE8 A0A2A4IU03 A0A3S2KXX3 Q16LT3 A0A023EMK4 A0A182H2Z1 A0A023EPW8 A0A023ENQ5 A0A023EMJ0 A0A023ESE8 B0W1E6 A0A1Q3FKK5 A0A1B6KDN1 A0A1Y1K6J6 A0A1B6MCQ8 A0A1J1HRI6 A0A1L8DK41 A0A1B6HCW8 A0A1L8DJV1 A0A1L8DJK5 A0A2J7QZJ0 A0A0J7L6K8 A0A1L8DKC0 A0A1B6HHR4 A0A1Y1K6D0 A0A1B6GIS6 D6X0E2 A0A1B6FPI6 W8AVJ1 A0A0A1X025 A0A1B0D6J3 U5EP47 A0A151XJT2 E2A973 A0A1B0CUZ7 A0A182IPX2 A0A067QFM7 A0A182RK40 W8ARL8 A0A3M0IVD9 A0A0K8W976 A0A0K8VE31 J3JUM3 A0A1S4GZL2 A0A182X5V5 A0A182IBI8 A0A182LRX5 C3UTC3 A0A182SHP8 A0A182NGQ4 A0A182WFF0 A0A182K5V7 A0A034W8M1 A0A0L0BXQ5 B4Q7A9 A0A195C3M0 A0A182P1L4 E9J782 A0A1A9W2K0 C1C4F6 A0A182YN77 A0A182QIV2 B4NXJ4 A0A3B3RV58 A0A1B0B299 A0A3B1IPS9 A0A3B3RV68 A0A158NJN4 A0A1L8EF34 A0A1L8EAQ7 B4KEY5 A0A1A9Y1P2 Q9VLH0 K1QBC6 A0A232FCT9 A0A1B0AAL1 A0A1A9VA65 D3TQ22 B4HYS5 A0A1V4J584 B3N7U7 F4WCE0 A0A195B5J4 A0A1W4VMH2 T1P8S2 A0A3B1J8E6 A0A0K8TPC7 A0A3N0YM71 A0A067QFC7 A0A1B6E2I7
PDB
5TCX
E-value=3.1488e-07,
Score=127
Ontologies
GO
PANTHER
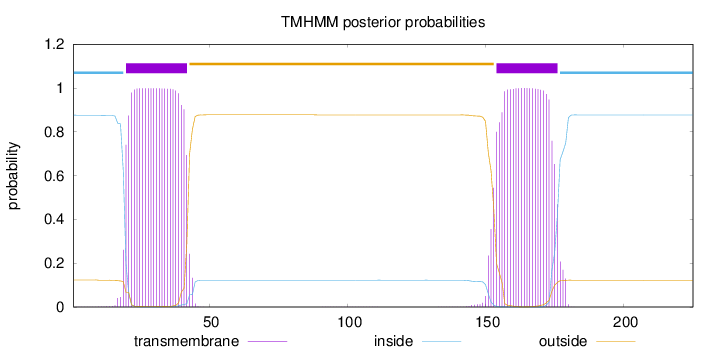
Topology
Subcellular location
Membrane
Length:
225
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.8516
Exp number, first 60 AAs:
22.80469
Total prob of N-in:
0.87616
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 153
TMhelix
154 - 176
inside
177 - 225
Population Genetic Test Statistics
Pi
190.219352
Theta
170.787815
Tajima's D
0.158485
CLR
1.953666
CSRT
0.417179141042948
Interpretation
Uncertain
