Gene
KWMTBOMO05912
Pre Gene Modal
BGIBMGA014593
Annotation
PREDICTED:_alpha-ketoglutarate-dependent_dioxygenase_alkB_homolog_4_[Amyelois_transitella]
Full name
DNA-(apurinic or apyrimidinic site) lyase
Location in the cell
Nuclear Reliability : 2.728
Sequence
CDS
ATGAAACCGAGACCTTGCGGCTGCAAAGGTTGTAGGACGTGTTTGATCTGTGAAACACAATACGGTGCTGCAGACCTGAAATTGCAGTTGGAACTAGACAAAGAAAAAAGTTACGTCTACTGTCCGTATTGCAACAAAGCCTGGAAAGGATGGGACATCAATTCTTACAAAGAACATCCGAACCATCACGGGGAACCGTTGAGTTATCGCGGCGTCTATATTCACTTGGACTTTATAAGCGAGTTGGAAGAGCAAATCTTAATCGATAACATAGACGAAATACCTTGGGATAACTCACAGAGTGGCAGAAGGAAACAGAACTATGGACCTAAAGTCAACTTCAAAAAGAAAAAAATCGTTTCCGGAAATTTCGAGGGTTTTCCAAAGTTTTCGAAAATGCTACAAGACAGATTTGCGACCATCGAAATGTTGGACGGTTTTGAAGTAATAGAACAGTGTTCATTGGAGTATGATCCCACTAAAGGTGCATCAATAGACCCACATATCGACGACTGCTGGGTGTGGGGTGAGAGAATCGTCACCGTTAACTTTCTGTCTGACTCAGTCCTCACAATGACACGTTTCAAAGGAGCGAAAACCAAATACAATTTATGCTCTGCTGAAAATTACCTTCCAGTGTTGGATTTAGAAGGAAAAATTATCAGAAATAATAAGAACCAAAGATCCTCTTTAGATATATGTAAACCTGTGACTGATGTGGATGTGATTGTCAGAATACCAATGCCAAGACGCTCATTACTAGTGCTGTATGGAGAATCGAGATATCACTGGGAGCATTGTGTACTCAGACAAGATATTTCAGCAAGGAGAGTATGTATTGCATATAGAGAGTTCACTCCCCCATACATGAATGGAGAACATCAGGAACTCGGTAAAGAAATACGAAGGAAAGGCAAAATGTTTTGGGATCATACTGAAAAATACCAAATAAATGTTAACTTAAATGAAAAGTGTGATATATTAAATGGTATATAA
Protein
MKPRPCGCKGCRTCLICETQYGAADLKLQLELDKEKSYVYCPYCNKAWKGWDINSYKEHPNHHGEPLSYRGVYIHLDFISELEEQILIDNIDEIPWDNSQSGRRKQNYGPKVNFKKKKIVSGNFEGFPKFSKMLQDRFATIEMLDGFEVIEQCSLEYDPTKGASIDPHIDDCWVWGERIVTVNFLSDSVLTMTRFKGAKTKYNLCSAENYLPVLDLEGKIIRNNKNQRSSLDICKPVTDVDVIVRIPMPRRSLLVLYGESRYHWEHCVLRQDISARRVCIAYREFTPPYMNGEHQELGKEIRRKGKMFWDHTEKYQINVNLNEKCDILNGI
Summary
Catalytic Activity
The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate.
Similarity
Belongs to the iron/ascorbate-dependent oxidoreductase family.
Belongs to the DNA repair enzymes AP/ExoA family.
Belongs to the DNA repair enzymes AP/ExoA family.
Uniprot
H9JYH3
S4PY47
A0A212F0E8
A0A0L7L5H6
A0A194Q428
A0A194QWH9
+ More
A0A3S2M9W8 A0A2A4IS25 A0A1E1W9W6 A0A2P8YLC1 A0A067RDL2 A0A195FR85 A0A2Z5U5Y9 F4WCD9 A0A0J7L5P2 A0A158NJQ8 A0A151XJR4 A0A3L8DDI7 A0A195EKL8 A0A195C5I8 A0A088AJM4 A0A1W4X0M0 A0A2A3E6V0 A0A0N0U3C5 E2A974 A0A1Y1JW78 A0A232FDA1 K7IWU2 D6WUI0 A0A154PJG9 A0A0C9RY19 Q16S01 A0A026WRZ8 W5JC07 A0A1B0D6X1 B0WX57 A0A182UMS7 A0A182L5X8 A0A182U7L1 A0A182XCJ0 J3JTX3 A0A182M8W1 A0A1Y9GLD5 A0A2M4AVC4 Q7PLZ9 A0A1Q3FDK2 A0A182GCY5 A0A1B6GT67 A0A1S3D6Z8 A0A1S3D5F8 A0A182PZD2 A0A1L8DFG9 A0A182F239 A0A1B6IHI2 A0A1B6KZD8 A0A1B6E1Q0 A0A182NUY2 A0A023EMG1 A0A182RRH2 U5EXP1 E9GAJ0 A0A2J7PJU2 A0A146MDD7 A0A0A9WWW8 A0A0L7QPN7 A0A0P5HFF2 A0A182K535 A0A146M525 A0A164SGQ8 A0A0P5HV87 A0A0P5WT63 A0A0P5RVZ7 A0A0P5MM12 A0A0P5W0Q5 A0A0P6BHP3 A0A0P5IK22 A0A0P6DBI6 A0A0P5TCW9 A0A0P5FRD7 A0A0N8C076 A0A0P6JPK7 A0A0P5LJQ3 A0A0P5IDE5 A0A0P5XPY3 A0A182PB10 A0A0K8VL64 B4NYM3 B3N8I5 A0A0Q9WJH2 Q9VL81 A0A0P4VS12 T1I7G8 W8CB18 Q8MR16 A0A1B0GIS2 B3MUZ6 B4JR58 B4HWA8 A0A1W4W9T5 A0A182Y5M2
A0A3S2M9W8 A0A2A4IS25 A0A1E1W9W6 A0A2P8YLC1 A0A067RDL2 A0A195FR85 A0A2Z5U5Y9 F4WCD9 A0A0J7L5P2 A0A158NJQ8 A0A151XJR4 A0A3L8DDI7 A0A195EKL8 A0A195C5I8 A0A088AJM4 A0A1W4X0M0 A0A2A3E6V0 A0A0N0U3C5 E2A974 A0A1Y1JW78 A0A232FDA1 K7IWU2 D6WUI0 A0A154PJG9 A0A0C9RY19 Q16S01 A0A026WRZ8 W5JC07 A0A1B0D6X1 B0WX57 A0A182UMS7 A0A182L5X8 A0A182U7L1 A0A182XCJ0 J3JTX3 A0A182M8W1 A0A1Y9GLD5 A0A2M4AVC4 Q7PLZ9 A0A1Q3FDK2 A0A182GCY5 A0A1B6GT67 A0A1S3D6Z8 A0A1S3D5F8 A0A182PZD2 A0A1L8DFG9 A0A182F239 A0A1B6IHI2 A0A1B6KZD8 A0A1B6E1Q0 A0A182NUY2 A0A023EMG1 A0A182RRH2 U5EXP1 E9GAJ0 A0A2J7PJU2 A0A146MDD7 A0A0A9WWW8 A0A0L7QPN7 A0A0P5HFF2 A0A182K535 A0A146M525 A0A164SGQ8 A0A0P5HV87 A0A0P5WT63 A0A0P5RVZ7 A0A0P5MM12 A0A0P5W0Q5 A0A0P6BHP3 A0A0P5IK22 A0A0P6DBI6 A0A0P5TCW9 A0A0P5FRD7 A0A0N8C076 A0A0P6JPK7 A0A0P5LJQ3 A0A0P5IDE5 A0A0P5XPY3 A0A182PB10 A0A0K8VL64 B4NYM3 B3N8I5 A0A0Q9WJH2 Q9VL81 A0A0P4VS12 T1I7G8 W8CB18 Q8MR16 A0A1B0GIS2 B3MUZ6 B4JR58 B4HWA8 A0A1W4W9T5 A0A182Y5M2
EC Number
4.2.99.18
Pubmed
19121390
23622113
22118469
26227816
26354079
29403074
+ More
24845553 26760975 21719571 21347285 30249741 20798317 28004739 28648823 20075255 18362917 19820115 17510324 24508170 20920257 23761445 20966253 22516182 12364791 14747013 17210077 26483478 24945155 21292972 26823975 25401762 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 24495485 25244985
24845553 26760975 21719571 21347285 30249741 20798317 28004739 28648823 20075255 18362917 19820115 17510324 24508170 20920257 23761445 20966253 22516182 12364791 14747013 17210077 26483478 24945155 21292972 26823975 25401762 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 24495485 25244985
EMBL
BABH01043572
GAIX01003918
JAA88642.1
AGBW02011105
OWR47203.1
JTDY01002796
+ More
KOB70707.1 KQ459472 KPJ00099.1 KQ461175 KPJ07926.1 RSAL01000003 RVE54772.1 NWSH01008489 PCG62541.1 GDQN01007254 GDQN01000787 JAT83800.1 JAT90267.1 PYGN01000512 PSN45038.1 KK852704 KDR18060.1 KQ981305 KYN42797.1 FX985796 BBA93683.1 GL888070 EGI68119.1 LBMM01000455 KMQ98297.1 ADTU01001951 KQ982052 KYQ60663.1 QOIP01000009 RLU18213.1 KQ978739 KYN28815.1 KQ978292 KYM95446.1 KZ288349 PBC27438.1 KQ435916 KOX68751.1 GL437711 EFN70059.1 GEZM01099011 JAV53574.1 NNAY01000426 OXU28490.1 AAZX01000576 KQ971357 EFA08384.1 KQ434924 KZC11624.1 GBYB01014170 JAG83937.1 CH477692 EAT37208.1 KK107119 EZA58825.1 ADMH02001945 ETN60379.1 AJVK01003764 DS232158 EDS36318.1 BT126683 AEE61645.1 AXCM01000826 APCN01004535 GGFK01011399 MBW44720.1 AAAB01008980 EAA14576.4 GFDL01009408 JAV25637.1 JXUM01055135 KQ561852 KXJ77336.1 GECZ01004153 JAS65616.1 AXCN02000752 GFDF01008896 JAV05188.1 GECU01021312 JAS86394.1 GEBQ01023160 JAT16817.1 GEDC01005440 GEDC01004910 JAS31858.1 JAS32388.1 GAPW01003090 JAC10508.1 GANO01000855 JAB59016.1 GL732537 EFX83520.1 NEVH01024944 PNF16603.1 GDHC01000798 JAQ17831.1 GBHO01031370 GBHO01005272 GBRD01011692 JAG12234.1 JAG38332.1 JAG54132.1 KQ414821 KOC60441.1 GDIQ01233761 JAK17964.1 GDHC01019654 GDHC01004517 JAP98974.1 JAQ14112.1 LRGB01002019 KZS09615.1 GDIQ01227440 JAK24285.1 GDIP01082110 JAM21605.1 GDIQ01095501 JAL56225.1 GDIQ01156230 JAK95495.1 GDIP01093414 JAM10301.1 GDIP01029010 JAM74705.1 GDIQ01212863 JAK38862.1 GDIQ01080512 JAN14225.1 GDIQ01093456 JAL58270.1 GDIQ01251496 JAK00229.1 GDIQ01127651 JAL24075.1 GDIQ01001645 JAN93092.1 GDIQ01193570 JAK58155.1 GDIQ01214948 JAK36777.1 GDIP01070361 JAM33354.1 GDHF01012682 JAI39632.1 CM000157 EDW88687.1 CH954177 EDV58408.1 CH940649 KRF81070.1 AE014134 AAF52815.1 ADV37012.1 GDKW01000722 JAI55873.1 ACPB03005592 GAMC01000849 JAC05707.1 AY122185 AAM52697.1 AJWK01015363 AJWK01015364 CH902624 EDV33061.1 CH916372 EDV99388.1 CH480818 EDW52303.1
KOB70707.1 KQ459472 KPJ00099.1 KQ461175 KPJ07926.1 RSAL01000003 RVE54772.1 NWSH01008489 PCG62541.1 GDQN01007254 GDQN01000787 JAT83800.1 JAT90267.1 PYGN01000512 PSN45038.1 KK852704 KDR18060.1 KQ981305 KYN42797.1 FX985796 BBA93683.1 GL888070 EGI68119.1 LBMM01000455 KMQ98297.1 ADTU01001951 KQ982052 KYQ60663.1 QOIP01000009 RLU18213.1 KQ978739 KYN28815.1 KQ978292 KYM95446.1 KZ288349 PBC27438.1 KQ435916 KOX68751.1 GL437711 EFN70059.1 GEZM01099011 JAV53574.1 NNAY01000426 OXU28490.1 AAZX01000576 KQ971357 EFA08384.1 KQ434924 KZC11624.1 GBYB01014170 JAG83937.1 CH477692 EAT37208.1 KK107119 EZA58825.1 ADMH02001945 ETN60379.1 AJVK01003764 DS232158 EDS36318.1 BT126683 AEE61645.1 AXCM01000826 APCN01004535 GGFK01011399 MBW44720.1 AAAB01008980 EAA14576.4 GFDL01009408 JAV25637.1 JXUM01055135 KQ561852 KXJ77336.1 GECZ01004153 JAS65616.1 AXCN02000752 GFDF01008896 JAV05188.1 GECU01021312 JAS86394.1 GEBQ01023160 JAT16817.1 GEDC01005440 GEDC01004910 JAS31858.1 JAS32388.1 GAPW01003090 JAC10508.1 GANO01000855 JAB59016.1 GL732537 EFX83520.1 NEVH01024944 PNF16603.1 GDHC01000798 JAQ17831.1 GBHO01031370 GBHO01005272 GBRD01011692 JAG12234.1 JAG38332.1 JAG54132.1 KQ414821 KOC60441.1 GDIQ01233761 JAK17964.1 GDHC01019654 GDHC01004517 JAP98974.1 JAQ14112.1 LRGB01002019 KZS09615.1 GDIQ01227440 JAK24285.1 GDIP01082110 JAM21605.1 GDIQ01095501 JAL56225.1 GDIQ01156230 JAK95495.1 GDIP01093414 JAM10301.1 GDIP01029010 JAM74705.1 GDIQ01212863 JAK38862.1 GDIQ01080512 JAN14225.1 GDIQ01093456 JAL58270.1 GDIQ01251496 JAK00229.1 GDIQ01127651 JAL24075.1 GDIQ01001645 JAN93092.1 GDIQ01193570 JAK58155.1 GDIQ01214948 JAK36777.1 GDIP01070361 JAM33354.1 GDHF01012682 JAI39632.1 CM000157 EDW88687.1 CH954177 EDV58408.1 CH940649 KRF81070.1 AE014134 AAF52815.1 ADV37012.1 GDKW01000722 JAI55873.1 ACPB03005592 GAMC01000849 JAC05707.1 AY122185 AAM52697.1 AJWK01015363 AJWK01015364 CH902624 EDV33061.1 CH916372 EDV99388.1 CH480818 EDW52303.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053268
UP000053240
UP000283053
+ More
UP000218220 UP000245037 UP000027135 UP000078541 UP000007755 UP000036403 UP000005205 UP000075809 UP000279307 UP000078492 UP000078542 UP000005203 UP000192223 UP000242457 UP000053105 UP000000311 UP000215335 UP000002358 UP000007266 UP000076502 UP000008820 UP000053097 UP000000673 UP000092462 UP000002320 UP000075903 UP000075882 UP000075902 UP000076407 UP000075883 UP000075840 UP000007062 UP000069940 UP000249989 UP000079169 UP000075886 UP000069272 UP000075884 UP000075900 UP000000305 UP000235965 UP000053825 UP000075881 UP000076858 UP000075885 UP000002282 UP000008711 UP000008792 UP000000803 UP000015103 UP000092461 UP000007801 UP000001070 UP000001292 UP000192221 UP000076408
UP000218220 UP000245037 UP000027135 UP000078541 UP000007755 UP000036403 UP000005205 UP000075809 UP000279307 UP000078492 UP000078542 UP000005203 UP000192223 UP000242457 UP000053105 UP000000311 UP000215335 UP000002358 UP000007266 UP000076502 UP000008820 UP000053097 UP000000673 UP000092462 UP000002320 UP000075903 UP000075882 UP000075902 UP000076407 UP000075883 UP000075840 UP000007062 UP000069940 UP000249989 UP000079169 UP000075886 UP000069272 UP000075884 UP000075900 UP000000305 UP000235965 UP000053825 UP000075881 UP000076858 UP000075885 UP000002282 UP000008711 UP000008792 UP000000803 UP000015103 UP000092461 UP000007801 UP000001070 UP000001292 UP000192221 UP000076408
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
H9JYH3
S4PY47
A0A212F0E8
A0A0L7L5H6
A0A194Q428
A0A194QWH9
+ More
A0A3S2M9W8 A0A2A4IS25 A0A1E1W9W6 A0A2P8YLC1 A0A067RDL2 A0A195FR85 A0A2Z5U5Y9 F4WCD9 A0A0J7L5P2 A0A158NJQ8 A0A151XJR4 A0A3L8DDI7 A0A195EKL8 A0A195C5I8 A0A088AJM4 A0A1W4X0M0 A0A2A3E6V0 A0A0N0U3C5 E2A974 A0A1Y1JW78 A0A232FDA1 K7IWU2 D6WUI0 A0A154PJG9 A0A0C9RY19 Q16S01 A0A026WRZ8 W5JC07 A0A1B0D6X1 B0WX57 A0A182UMS7 A0A182L5X8 A0A182U7L1 A0A182XCJ0 J3JTX3 A0A182M8W1 A0A1Y9GLD5 A0A2M4AVC4 Q7PLZ9 A0A1Q3FDK2 A0A182GCY5 A0A1B6GT67 A0A1S3D6Z8 A0A1S3D5F8 A0A182PZD2 A0A1L8DFG9 A0A182F239 A0A1B6IHI2 A0A1B6KZD8 A0A1B6E1Q0 A0A182NUY2 A0A023EMG1 A0A182RRH2 U5EXP1 E9GAJ0 A0A2J7PJU2 A0A146MDD7 A0A0A9WWW8 A0A0L7QPN7 A0A0P5HFF2 A0A182K535 A0A146M525 A0A164SGQ8 A0A0P5HV87 A0A0P5WT63 A0A0P5RVZ7 A0A0P5MM12 A0A0P5W0Q5 A0A0P6BHP3 A0A0P5IK22 A0A0P6DBI6 A0A0P5TCW9 A0A0P5FRD7 A0A0N8C076 A0A0P6JPK7 A0A0P5LJQ3 A0A0P5IDE5 A0A0P5XPY3 A0A182PB10 A0A0K8VL64 B4NYM3 B3N8I5 A0A0Q9WJH2 Q9VL81 A0A0P4VS12 T1I7G8 W8CB18 Q8MR16 A0A1B0GIS2 B3MUZ6 B4JR58 B4HWA8 A0A1W4W9T5 A0A182Y5M2
A0A3S2M9W8 A0A2A4IS25 A0A1E1W9W6 A0A2P8YLC1 A0A067RDL2 A0A195FR85 A0A2Z5U5Y9 F4WCD9 A0A0J7L5P2 A0A158NJQ8 A0A151XJR4 A0A3L8DDI7 A0A195EKL8 A0A195C5I8 A0A088AJM4 A0A1W4X0M0 A0A2A3E6V0 A0A0N0U3C5 E2A974 A0A1Y1JW78 A0A232FDA1 K7IWU2 D6WUI0 A0A154PJG9 A0A0C9RY19 Q16S01 A0A026WRZ8 W5JC07 A0A1B0D6X1 B0WX57 A0A182UMS7 A0A182L5X8 A0A182U7L1 A0A182XCJ0 J3JTX3 A0A182M8W1 A0A1Y9GLD5 A0A2M4AVC4 Q7PLZ9 A0A1Q3FDK2 A0A182GCY5 A0A1B6GT67 A0A1S3D6Z8 A0A1S3D5F8 A0A182PZD2 A0A1L8DFG9 A0A182F239 A0A1B6IHI2 A0A1B6KZD8 A0A1B6E1Q0 A0A182NUY2 A0A023EMG1 A0A182RRH2 U5EXP1 E9GAJ0 A0A2J7PJU2 A0A146MDD7 A0A0A9WWW8 A0A0L7QPN7 A0A0P5HFF2 A0A182K535 A0A146M525 A0A164SGQ8 A0A0P5HV87 A0A0P5WT63 A0A0P5RVZ7 A0A0P5MM12 A0A0P5W0Q5 A0A0P6BHP3 A0A0P5IK22 A0A0P6DBI6 A0A0P5TCW9 A0A0P5FRD7 A0A0N8C076 A0A0P6JPK7 A0A0P5LJQ3 A0A0P5IDE5 A0A0P5XPY3 A0A182PB10 A0A0K8VL64 B4NYM3 B3N8I5 A0A0Q9WJH2 Q9VL81 A0A0P4VS12 T1I7G8 W8CB18 Q8MR16 A0A1B0GIS2 B3MUZ6 B4JR58 B4HWA8 A0A1W4W9T5 A0A182Y5M2
PDB
3THT
E-value=0.0399674,
Score=85
Ontologies
GO
Topology
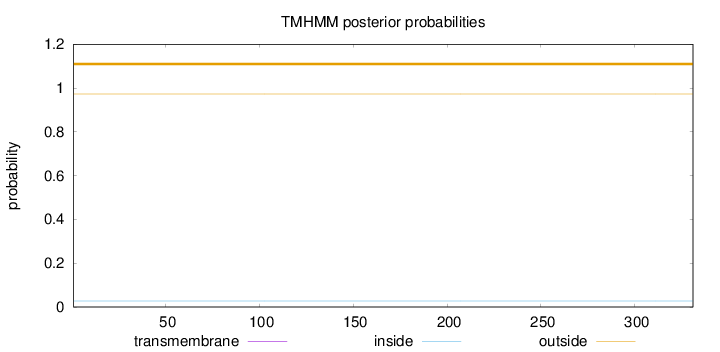
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00054
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02734
outside
1 - 331
Population Genetic Test Statistics
Pi
139.717756
Theta
133.092029
Tajima's D
0.982939
CLR
3.049485
CSRT
0.655517224138793
Interpretation
Uncertain
