Gene
KWMTBOMO05911
Pre Gene Modal
BGIBMGA002785
Annotation
PREDICTED:_echinoderm_microtubule-associated_protein-like_CG42247_[Amyelois_transitella]
Full name
Echinoderm microtubule-associated protein-like CG42247
Location in the cell
Mitochondrial Reliability : 1.018 Nuclear Reliability : 1.479
Sequence
CDS
ATGTCGCGTCGCGTACACCGCCCGCGGATACAAGCCCGTCTCCGCCCGCAGGCCGCCCCGGAGCCCGCACCGCCACCGGAACCGCCCCAACCTGAGCCCGACCATCCGCCTTCCGAAGAGGAACGAGAACCAGGCGACTGGCGAGCCTCTCGGCCATCCTCCCCTCCACAGCGCCCTGATGACATCTCAGAAGGCTCGGAACCTCCACGACCGATCAGAACAGAACCCGCCAGTCGCTATGCCAACCTCAACTACTGGAAAGCGAGAAGGGTCACGTTCTACCGTAACGGAGATCCGTTCCATCCTGGCGTCGAATTCCGATTCAAGCCGGGTCGTGATTTGGCGTCTATTGATGCGTTACTGGACCGATTGTCTGAGAGACTAGAATTACCTAGAGGAGCGAGGTTTATCTTCGGGATGGACGGAGACCGGAAGTACAGGCTGGAAGAGCTGGAAGACGGAGCATCTTACGTCGTGTCTTCCTACAAAACTTTCAAGGCCGCCAGCTACGGTAAAAAGAACGGTATGTGGTACGGTGGAACTGGTACCAGCGGGTGGTCGAGGTCCGGTACTCGCAAGCAATCGGTTGCGGAGTCCGATGCACCTCCTCCCGGAGGTGGAAAGCCGGCTAGTGGTCGTGTCATCAGAATCATCAACAACATGGACCACTCTATACAGTGTCGCGTTTTACTGAACTTACGGACAACGCAACCTTTCGAAGAGGTATTAGAGGATCTGGGTCAAGTACTCAAAATGAGCGGGGCAAAGCGAATGTACACGATAACCGGTCAAGAGGTAAGAAGTTTCTCTCAACTCCGGAACGAATTTGCAGACGTGGAAACATTTTACTTGGGACCTGCGATGCTTCCACCAGCTTTGAGTCCCGGAGTCAGCGCTCCACTGCCTATCGAGTCACCAATTAGAAGGTCTCGATCGAGAGCCAATGTGGCGACAGCACCGATGATTGATGACACGCGAAACGGTGGGCGACGAGCCAGGAGCAAGAGCAGACCGCGGGTTTTATACGCGCCCGAAGGCGAGATTGTTAGAAATTCAGATTACACTCTCCTTGAAGTTCTGAAGGAAGAACCTATACGCGTGACTATTCGTGGCTTACGAAGAACATTCTACCCGCCCATACATCATGCGCCTGTCGACAACTCTCCGCCCGACAAGAAATTACAACTAGATTGGGTATATGGGTATCGAGGTTTTGATTCCCGTCGAAACTTATGGGTGTTGCCGACTGGAGAACTGCTGTACTACGTGGCGGCTGTGGCCGTCATGTACGACAGAGATGAGGAGTCTCAAAGACATTATACCGGCCACACTGAGGATATACAGTGCATGGAGCTCCATCCGTCCCGGGAGCTTGTGGCGAGCGGACAACGGGCCGGCCGGGGACGGCGCGCTCAGGCGCACGTCCGCATCTGGAGCACCGACACATTGCAGACCCTGCACGTGTTCGGCATGGCCGAATTCGAAGCCGGTGTCTCGGCTGTGGCATTCTCGCAATTAAATGGCGGTAGTTACGTCCTTGCTGTGGATGCTGGTCGCGAGAGCATACTCTCCGTGTGGCAGTGGCAGTGGGGCCATCTACTCGGAAAAGTCGCTACTTTGCAAGAAGAGCTAACCGGCGCTGCTTTTCATCCTTTAGACGACAACTTACTCATAACACACGGAAAAGGCCATCTCGCGTTCTGGAATAGAAGGAAAGATGGATTCTTCGAAAGAACCGATATCGTCAAACCGCCTTCTCGCACGCATGTGACCGCCCTGCAGTTCGAGCAGGACGGCGACGTGGTGACGGCAGATAGCGATGGGTTCATTACGATTTACAGCGTCGATAGCGACGGTGCTTACTTCGTGCGAATGGAGTTTGAGGCTCACATCAAAGGCATAAGTTCCCTCATCATGCTGTCTGAGGGGACATTAATATCTGGAGGCGAGAAGGACAGAAAAATTGCCGCCTGGGACTCTTTGCAGAATTACAAAAGAATTACTGAGACGAAGTTACCAGAAACTGCTGGTGGGATCCGCAGTATCTACCCACAGCGGCCCGGTCGTAACGATGGTAATCTGTACGTCGGTACGACCAGAAACAATATAATGGAGGGATCTTTACAGAGGCGATTTAATCAAATTGTCTTCGGACACCACAAGCCGCTGCTGGGTGTCGCGGTACACCCTGACGACGAGATGTTCGCGACAGCCGGTCACGACAAGAACATTGCTCTGTGGAAAGGGCACAAGCTTGTTTTCGCAACTCAGGTGGGATACGAGTGCGTGTCGCTGGCGTGGCACCCCGGGGGCGGCGCGCTCGCGGCCGGCAGCACTGAGGGTCACCTGGTTGTGCTCAACGCCGACGCCGGAGCGCATGTCGCAACACTCAGAGTCTGCGGTTCGCCGCTCAATTGCCTGCAATATAACACCGCTGGAGATTTCCTCGCCATTGGATCTCAGAACGGCAGTATCTACCTATTCAGGGTGTCTCGAGACGGCTTCTCTTACAAGAAGTCGAATAAGATCCGAGGCGCTCAACCTCTGATGCAGTTGGACTGGAGTCTCGACGGAAACTTCTTGCAGACTGTGACCGCGGATTACGATTTGGCTTTCTGGGATATAAAATCGCTATCTCCAGAGAAAAGCCCGATAGGCATGAAAGATGTCAAGTGGTCCACGTTTAACTCAACTGTCGGATTCCTTGTCTCGGGTATGTGGAACAATCGCTTCTACCCAATGACATCCTTGATTTCGTCATCGAGTCGCTCGGCGGCCCACGATCTTCTGATCAGCGGTGACTACGATGGATACCTACGACTGTTCAGGTATCCATGCGCCAGCCCTAAGGCTGAATACAACGAGGTGAAAGTATATAGTGGCGCCATCCACAACGCGAGGTTCTTATTCAACGACCGCTGCGTGATCAGCGCCGGAGGTCACGACGCCGCTCTCATGCTGTGGGAGCTGGTGGAGGAATAG
Protein
MSRRVHRPRIQARLRPQAAPEPAPPPEPPQPEPDHPPSEEEREPGDWRASRPSSPPQRPDDISEGSEPPRPIRTEPASRYANLNYWKARRVTFYRNGDPFHPGVEFRFKPGRDLASIDALLDRLSERLELPRGARFIFGMDGDRKYRLEELEDGASYVVSSYKTFKAASYGKKNGMWYGGTGTSGWSRSGTRKQSVAESDAPPPGGGKPASGRVIRIINNMDHSIQCRVLLNLRTTQPFEEVLEDLGQVLKMSGAKRMYTITGQEVRSFSQLRNEFADVETFYLGPAMLPPALSPGVSAPLPIESPIRRSRSRANVATAPMIDDTRNGGRRARSKSRPRVLYAPEGEIVRNSDYTLLEVLKEEPIRVTIRGLRRTFYPPIHHAPVDNSPPDKKLQLDWVYGYRGFDSRRNLWVLPTGELLYYVAAVAVMYDRDEESQRHYTGHTEDIQCMELHPSRELVASGQRAGRGRRAQAHVRIWSTDTLQTLHVFGMAEFEAGVSAVAFSQLNGGSYVLAVDAGRESILSVWQWQWGHLLGKVATLQEELTGAAFHPLDDNLLITHGKGHLAFWNRRKDGFFERTDIVKPPSRTHVTALQFEQDGDVVTADSDGFITIYSVDSDGAYFVRMEFEAHIKGISSLIMLSEGTLISGGEKDRKIAAWDSLQNYKRITETKLPETAGGIRSIYPQRPGRNDGNLYVGTTRNNIMEGSLQRRFNQIVFGHHKPLLGVAVHPDDEMFATAGHDKNIALWKGHKLVFATQVGYECVSLAWHPGGGALAAGSTEGHLVVLNADAGAHVATLRVCGSPLNCLQYNTAGDFLAIGSQNGSIYLFRVSRDGFSYKKSNKIRGAQPLMQLDWSLDGNFLQTVTADYDLAFWDIKSLSPEKSPIGMKDVKWSTFNSTVGFLVSGMWNNRFYPMTSLISSSSRSAAHDLLISGDYDGYLRLFRYPCASPKAEYNEVKVYSGAIHNARFLFNDRCVISAGGHDAALMLWELVEE
Summary
Description
May modify the assembly dynamics of microtubules, such that microtubules are slightly longer, but more dynamic.
Similarity
Belongs to the WD repeat EMAP family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Reference proteome
Repeat
WD repeat
Feature
chain Echinoderm microtubule-associated protein-like CG42247
splice variant In isoform D.
splice variant In isoform D.
Uniprot
A0A2H1WHD9
A0A212F0I0
A0A194QSG6
A0A1W4WFE9
A0A194Q3I1
T1HQX8
+ More
A0A195AZ74 K7IT63 E2BZ47 A0A026W4T0 A0A0M3QWA6 Q2M179 A0A1W4UL58 A0A0J9UL53 Q9VUI3 B4PII0 B3NHJ7 M9PF94 B3MAV1 A0A3B0JY66 A0A1J1HYG1 A0A182IPW8 A0A158NZ51 Q7QFI4 D6X2P5 A0A2H8TVI9 A0A2S2NPA7 J9JQL4 A0A0C9Q1W1 A0A0M9A361 A0A195D0N2 A0A088A158 A0A195F7C6 A0A151WWX9 A0A151IS42 Q16H05 Q16LA1 A0A1S4FX01 B4LFM2 B4IY12 B4QKG9 A0A1W4UL98 B4KXN1 M9PFN2 A0A3B0JQV1 B4NM07 A0A1I8Q5R6 A0A0L0C8B5 B0WSV5 A0A182MZJ0 A0A182PFV2 A0A182W4I9 A0A182XE01 A0A182M0F5 A0A182UX81 A0A182UAS9 A0A182Y7S4 A0A1A9WCU1 A0A1A9Z8T2 A0A2A3ELV4 Q9VUI3-2 B4HHU6 A0A1B0CH65 A0A3F2YSR1 A0A3F2YYW5 A0A182FMA9 A0A1B0FI13 A0A336L3M0 A0A182R9H2 A0A336L5V7 A0A2S2R6X4 A0A1I8MCA3 U4UA20 A0A1B0D286 A0A336L6C9 E2AAR9 B4GUI1 A0A182G3U9 E0VAV1 N6TEJ2 A0A1A9Y9W5 A0A1B0BJX9 E9JD10 A0A3Q0J3X8
A0A195AZ74 K7IT63 E2BZ47 A0A026W4T0 A0A0M3QWA6 Q2M179 A0A1W4UL58 A0A0J9UL53 Q9VUI3 B4PII0 B3NHJ7 M9PF94 B3MAV1 A0A3B0JY66 A0A1J1HYG1 A0A182IPW8 A0A158NZ51 Q7QFI4 D6X2P5 A0A2H8TVI9 A0A2S2NPA7 J9JQL4 A0A0C9Q1W1 A0A0M9A361 A0A195D0N2 A0A088A158 A0A195F7C6 A0A151WWX9 A0A151IS42 Q16H05 Q16LA1 A0A1S4FX01 B4LFM2 B4IY12 B4QKG9 A0A1W4UL98 B4KXN1 M9PFN2 A0A3B0JQV1 B4NM07 A0A1I8Q5R6 A0A0L0C8B5 B0WSV5 A0A182MZJ0 A0A182PFV2 A0A182W4I9 A0A182XE01 A0A182M0F5 A0A182UX81 A0A182UAS9 A0A182Y7S4 A0A1A9WCU1 A0A1A9Z8T2 A0A2A3ELV4 Q9VUI3-2 B4HHU6 A0A1B0CH65 A0A3F2YSR1 A0A3F2YYW5 A0A182FMA9 A0A1B0FI13 A0A336L3M0 A0A182R9H2 A0A336L5V7 A0A2S2R6X4 A0A1I8MCA3 U4UA20 A0A1B0D286 A0A336L6C9 E2AAR9 B4GUI1 A0A182G3U9 E0VAV1 N6TEJ2 A0A1A9Y9W5 A0A1B0BJX9 E9JD10 A0A3Q0J3X8
Pubmed
EMBL
ODYU01008695
SOQ52490.1
AGBW02011105
OWR47204.1
KQ461175
KPJ07925.1
+ More
KQ459472 KPJ00098.1 ACPB03023001 ACPB03023002 ACPB03023003 ACPB03023004 KQ976701 KYM77259.1 AAZX01003328 GL451555 EFN79049.1 KK107419 EZA51062.1 CP012525 ALC43813.1 CH379069 EAL30696.2 CM002912 KMY99635.1 AE014296 BT001524 BT030411 AF208486 CM000159 EDW94537.2 CH954178 EDV51792.2 AGB94543.1 CH902618 EDV39185.2 OUUW01000002 SPP76038.1 CVRI01000020 CRK91169.1 ADTU01003814 ADTU01003815 ADTU01003816 ADTU01003817 AAAB01008846 EAA06456.3 KQ971372 EFA10279.1 GFXV01006480 MBW18285.1 GGMR01006348 MBY18967.1 ABLF02032629 ABLF02032634 ABLF02032636 ABLF02032643 GBYB01007968 JAG77735.1 KQ435776 KOX74879.1 KQ977004 KYN06475.1 KQ981768 KYN35974.1 KQ982684 KYQ52348.1 KQ981093 KYN09533.1 CH478218 EAT33525.1 CH477913 EAT35090.1 CH940647 EDW70340.2 CH916366 EDV97555.1 CM000363 EDX10480.1 CH933809 EDW19738.1 AGB94544.1 SPP76039.1 CH964278 EDW85375.2 JRES01000760 KNC28511.1 DS232077 EDS34083.1 AXCM01000723 AXCM01000724 AXCM01000725 KZ288215 PBC32687.1 CH480815 EDW41511.1 AJWK01012074 APCN01005676 CCAG010002770 CCAG010002771 UFQS01001714 UFQT01001714 SSX11941.1 SSX31488.1 SSX11940.1 SSX31487.1 GGMS01016583 MBY85786.1 KB631865 ERL86795.1 AJVK01022611 AJVK01022612 AJVK01022613 SSX11942.1 SSX31489.1 GL438158 EFN69469.1 CH479191 EDW26264.1 JXUM01142273 JXUM01142274 JXUM01142275 KQ569392 KXJ68640.1 DS235016 EEB10507.1 APGK01040998 APGK01040999 APGK01041000 APGK01041001 KB740986 ENN76153.1 JXJN01015692 GL771866 EFZ09341.1
KQ459472 KPJ00098.1 ACPB03023001 ACPB03023002 ACPB03023003 ACPB03023004 KQ976701 KYM77259.1 AAZX01003328 GL451555 EFN79049.1 KK107419 EZA51062.1 CP012525 ALC43813.1 CH379069 EAL30696.2 CM002912 KMY99635.1 AE014296 BT001524 BT030411 AF208486 CM000159 EDW94537.2 CH954178 EDV51792.2 AGB94543.1 CH902618 EDV39185.2 OUUW01000002 SPP76038.1 CVRI01000020 CRK91169.1 ADTU01003814 ADTU01003815 ADTU01003816 ADTU01003817 AAAB01008846 EAA06456.3 KQ971372 EFA10279.1 GFXV01006480 MBW18285.1 GGMR01006348 MBY18967.1 ABLF02032629 ABLF02032634 ABLF02032636 ABLF02032643 GBYB01007968 JAG77735.1 KQ435776 KOX74879.1 KQ977004 KYN06475.1 KQ981768 KYN35974.1 KQ982684 KYQ52348.1 KQ981093 KYN09533.1 CH478218 EAT33525.1 CH477913 EAT35090.1 CH940647 EDW70340.2 CH916366 EDV97555.1 CM000363 EDX10480.1 CH933809 EDW19738.1 AGB94544.1 SPP76039.1 CH964278 EDW85375.2 JRES01000760 KNC28511.1 DS232077 EDS34083.1 AXCM01000723 AXCM01000724 AXCM01000725 KZ288215 PBC32687.1 CH480815 EDW41511.1 AJWK01012074 APCN01005676 CCAG010002770 CCAG010002771 UFQS01001714 UFQT01001714 SSX11941.1 SSX31488.1 SSX11940.1 SSX31487.1 GGMS01016583 MBY85786.1 KB631865 ERL86795.1 AJVK01022611 AJVK01022612 AJVK01022613 SSX11942.1 SSX31489.1 GL438158 EFN69469.1 CH479191 EDW26264.1 JXUM01142273 JXUM01142274 JXUM01142275 KQ569392 KXJ68640.1 DS235016 EEB10507.1 APGK01040998 APGK01040999 APGK01041000 APGK01041001 KB740986 ENN76153.1 JXJN01015692 GL771866 EFZ09341.1
Proteomes
UP000007151
UP000053240
UP000192223
UP000053268
UP000015103
UP000078540
+ More
UP000002358 UP000008237 UP000053097 UP000092553 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000007801 UP000268350 UP000183832 UP000075880 UP000005205 UP000007062 UP000007266 UP000007819 UP000053105 UP000078542 UP000005203 UP000078541 UP000075809 UP000078492 UP000008820 UP000008792 UP000001070 UP000000304 UP000009192 UP000007798 UP000095300 UP000037069 UP000002320 UP000075884 UP000075885 UP000075920 UP000076407 UP000075883 UP000075903 UP000075902 UP000076408 UP000091820 UP000092445 UP000242457 UP000001292 UP000092461 UP000075840 UP000069272 UP000092444 UP000075900 UP000095301 UP000030742 UP000092462 UP000000311 UP000008744 UP000069940 UP000249989 UP000009046 UP000019118 UP000092443 UP000092460 UP000079169
UP000002358 UP000008237 UP000053097 UP000092553 UP000001819 UP000192221 UP000000803 UP000002282 UP000008711 UP000007801 UP000268350 UP000183832 UP000075880 UP000005205 UP000007062 UP000007266 UP000007819 UP000053105 UP000078542 UP000005203 UP000078541 UP000075809 UP000078492 UP000008820 UP000008792 UP000001070 UP000000304 UP000009192 UP000007798 UP000095300 UP000037069 UP000002320 UP000075884 UP000075885 UP000075920 UP000076407 UP000075883 UP000075903 UP000075902 UP000076408 UP000091820 UP000092445 UP000242457 UP000001292 UP000092461 UP000075840 UP000069272 UP000092444 UP000075900 UP000095301 UP000030742 UP000092462 UP000000311 UP000008744 UP000069940 UP000249989 UP000009046 UP000019118 UP000092443 UP000092460 UP000079169
PRIDE
Interpro
IPR019775
WD40_repeat_CS
+ More
IPR036572 Doublecortin_dom_sf
IPR017986 WD40_repeat_dom
IPR005108 HELP
IPR001680 WD40_repeat
IPR003533 Doublecortin_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR011041 Quinoprot_gluc/sorb_DH
IPR036572 Doublecortin_dom_sf
IPR017986 WD40_repeat_dom
IPR005108 HELP
IPR001680 WD40_repeat
IPR003533 Doublecortin_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR011041 Quinoprot_gluc/sorb_DH
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2H1WHD9
A0A212F0I0
A0A194QSG6
A0A1W4WFE9
A0A194Q3I1
T1HQX8
+ More
A0A195AZ74 K7IT63 E2BZ47 A0A026W4T0 A0A0M3QWA6 Q2M179 A0A1W4UL58 A0A0J9UL53 Q9VUI3 B4PII0 B3NHJ7 M9PF94 B3MAV1 A0A3B0JY66 A0A1J1HYG1 A0A182IPW8 A0A158NZ51 Q7QFI4 D6X2P5 A0A2H8TVI9 A0A2S2NPA7 J9JQL4 A0A0C9Q1W1 A0A0M9A361 A0A195D0N2 A0A088A158 A0A195F7C6 A0A151WWX9 A0A151IS42 Q16H05 Q16LA1 A0A1S4FX01 B4LFM2 B4IY12 B4QKG9 A0A1W4UL98 B4KXN1 M9PFN2 A0A3B0JQV1 B4NM07 A0A1I8Q5R6 A0A0L0C8B5 B0WSV5 A0A182MZJ0 A0A182PFV2 A0A182W4I9 A0A182XE01 A0A182M0F5 A0A182UX81 A0A182UAS9 A0A182Y7S4 A0A1A9WCU1 A0A1A9Z8T2 A0A2A3ELV4 Q9VUI3-2 B4HHU6 A0A1B0CH65 A0A3F2YSR1 A0A3F2YYW5 A0A182FMA9 A0A1B0FI13 A0A336L3M0 A0A182R9H2 A0A336L5V7 A0A2S2R6X4 A0A1I8MCA3 U4UA20 A0A1B0D286 A0A336L6C9 E2AAR9 B4GUI1 A0A182G3U9 E0VAV1 N6TEJ2 A0A1A9Y9W5 A0A1B0BJX9 E9JD10 A0A3Q0J3X8
A0A195AZ74 K7IT63 E2BZ47 A0A026W4T0 A0A0M3QWA6 Q2M179 A0A1W4UL58 A0A0J9UL53 Q9VUI3 B4PII0 B3NHJ7 M9PF94 B3MAV1 A0A3B0JY66 A0A1J1HYG1 A0A182IPW8 A0A158NZ51 Q7QFI4 D6X2P5 A0A2H8TVI9 A0A2S2NPA7 J9JQL4 A0A0C9Q1W1 A0A0M9A361 A0A195D0N2 A0A088A158 A0A195F7C6 A0A151WWX9 A0A151IS42 Q16H05 Q16LA1 A0A1S4FX01 B4LFM2 B4IY12 B4QKG9 A0A1W4UL98 B4KXN1 M9PFN2 A0A3B0JQV1 B4NM07 A0A1I8Q5R6 A0A0L0C8B5 B0WSV5 A0A182MZJ0 A0A182PFV2 A0A182W4I9 A0A182XE01 A0A182M0F5 A0A182UX81 A0A182UAS9 A0A182Y7S4 A0A1A9WCU1 A0A1A9Z8T2 A0A2A3ELV4 Q9VUI3-2 B4HHU6 A0A1B0CH65 A0A3F2YSR1 A0A3F2YYW5 A0A182FMA9 A0A1B0FI13 A0A336L3M0 A0A182R9H2 A0A336L5V7 A0A2S2R6X4 A0A1I8MCA3 U4UA20 A0A1B0D286 A0A336L6C9 E2AAR9 B4GUI1 A0A182G3U9 E0VAV1 N6TEJ2 A0A1A9Y9W5 A0A1B0BJX9 E9JD10 A0A3Q0J3X8
PDB
4CI8
E-value=1.46551e-111,
Score=1034
Ontologies
GO
Topology
Subcellular location
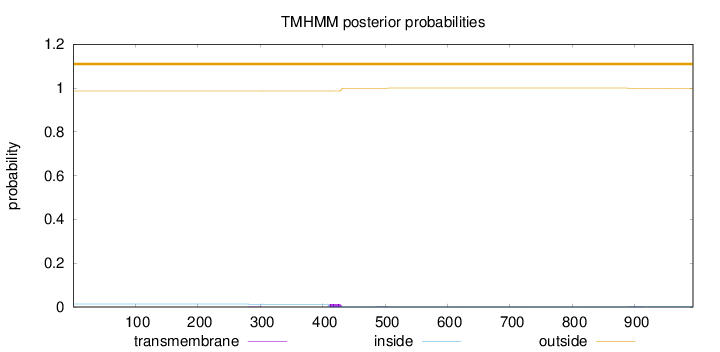
Length:
993
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.308180000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01326
outside
1 - 993
Population Genetic Test Statistics
Pi
224.974549
Theta
172.38944
Tajima's D
1.05987
CLR
0.370893
CSRT
0.677566121693915
Interpretation
Uncertain
