Gene
KWMTBOMO05909
Pre Gene Modal
BGIBMGA002921
Annotation
PREDICTED:_phosphatidylinositol_4?5-bisphosphate_3-kinase_catalytic_subunit_delta_isoform_[Papilio_xuthus]
Full name
Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit
+ More
Phosphatidylinositol 3-kinase catalytic subunit type 3
Phosphatidylinositol 3-kinase catalytic subunit type 3
Location in the cell
Nuclear Reliability : 3.167
Sequence
CDS
ATGGTGCCTGCGCCGAGTTGTCCCAATACGTGGGACTACTGGATGTCCCCGCAGTCCGATTGCGTAGAACTTACGTGCCTGCTCCCGAATTCAATGTACATACCCCTACGAAAAAGTTGGGATGCAACTTTTGAGGATGTCAAAGAGGATCTTTGGGACAGAGCAGTCAATTATCCTTTATTCGGAATGTTGCTTGAAATGAACAAATATATTTTTCAATTCGTAAACTCATTAGCTGTCACAGAAGAATTAGATGATGAAAGAAAACGTATATGTGACATCAAACCTGTATGTGGTGTGCTTATTATAGTTAAAAGGTCAGATGGTAGACCTGCCGAAGACTGCCTTAACAGAGACATCACACATTTGATAGGAAAATCCCTAAAAGTATTTGATAGTCTAAAAAATTCAGAAGTGAACGATTATCGAATGCGAATGCGCTATCTGGCCGAGGAGAGTTTGTTGAAACGGACGCACAGCACCACGCTCGAACGTCTCCGGTATCAGTGTCCGCTGCGGCTGGCCGATCAGCCCGAGATCCCGACAACACTGAAGAGTCATCTTAATGACCAGCGTTTCATACTCGTTACCAAAGTTATGGAGATAACCTCGTACTCATCAAACGTGCCGGCCACGTTGACTCCTCAGCAACACATCGAAATCGTGCTTCGAAAGTTGAACAACTCCATGAATTTGAGGTCGGAACATCCTCACAACTACGTTCTCAAAGTGTGCGGCCGTGAGGAGTACCTATTCGGGGATTATCCTTTGATACAGTTTCTATACATACAAGAGATGCTGGCCGTAGACGGAGTGCCTCAAGTTATGACCGTCAGTATTGATAAATTGCGTTTATCCTTGGATCGTGAGATACCATATATGCCGGAACGCAGAAGACTTACATCGGAACATTCGAACACTATGCGAAGAAAGAAAAACGAATCGTCTTGGAACATTCAGAAGAATTACACTTGCCGCGTGCAAAGCGCTTGCGGACTCAATGTGGACCCAAGCAGAGTGGTCGAGGTTGTCTGTTTAGGCGGGATATTTCACGGCGGCAAGCCGCTATGCGAAGTACAAAAGACAAAAGTTGCTGTAATATCGAAAGAAGGTGTTGCCACATGGGACGAAGAGTTGACATTTCCGATAAAAGTTCACAACTTACCGCGAATGGCGAGACTGTGTTTCGGTATTTACGAACTATCAAAGATTAAATCGAAATGGAGAGGAAAGGATAAGGAGCCGGTGAATCGATTGACGTGGGCCAACACAATGGTGTTCGATTACAAGGAGCAGCTGCGCACGGACGGCGTGCGGCTCGCGATGTGGACGCACGTGGCGGACGACACGCAGGGCGACGACATGGTGCTGCACCCGCTCGGCACCGTCGTCGCCAACCCCACGCCCGACTCCGCCGTGCTGCAGGTCATGTTCTCCAATTACGACTGCCAATATCCTATCCTATTCCCGAAGCAAGAGACAGTGATGGCTTACTCGGACCACATCGAGAGCGTCTCCCCGGACTTGATGGACAGACTGTGCAGAGACTTTGAACGCTTACGTATAATAGCCGAGAAAGATCCTATGTATGAAATGCACGAGCAAGAGAAAAAAGATATTTGGGCTTCGAGAGAACGCTTTCGAGTGGAAACGCCCGAGCTACTGCCAAAGTTATTGTCATGCGTTGAGTGGGGTGAACGAACTCAGGCCTCCAGCGTGCCCAGGATGTTGGACGAGTGGCCTCTGTTGCGGGTGGAGTCTGCCTTAGAGCTGTTGGACTACGCGTACGCTGATGCAGCAGTCAGGAGTTTTGCTGTCAAATGTTTGGAAAAAATTAGTGATAACGATCTCATGCTGTACCTCCTCCAGTTGGTTCAAGCCCTGAAGCACGAGTCCTACCTCATGTGTGACCTGTCCGTGTTCCTCCTGCAGCGAGCCTTCAAGAATATGACCATTGGACACTATTTATTTTGGCATTTAAGGTCGGAGATGCACGTGCCGTCCGTGTCCGTGCGCTTCCGGCTGATGCTCGAGGCGTACTGCCGCGGCTCGCAAGAGCACATCAAGATACTCAAGCAACAGATCACGTGTCTGGAGAAACTTAAATGGGTGAATCAGTGTGTTAAAGAGAAAAAGGAGATATCTAAAGCCCGAATCGCGTTACACCAGCACTTACAAGAGAAACACTGCGTCGACGCTCTCTGCGAGGGATTCGTGTCGCCGCTCAACCCTAGCTTCAAGTGTAAAAGAATCAAGCCTGAAGGTTGTCGAGTCATGGACAGTAAAATGCGTCCTCTGATATTGAATTTTGAGAATTTCGACCCGACCGGCAACGATATCCGGATAATATTGAAAATAGGAGACGATCTCAGACAAGATATGTTCACGCTACAGATGCTGAGGATAATGGATAGATTATGGAAAAATCATGGATACGACTTCAGACTGAACGCGTACAACTGCATATCGATCGAGTATTCTGTCGGCATGATCGAGGTGGTGGAGGACGCGGAGACCGTCGCCAACATACAGAAGCAGAACGCACCCTTCAACGCGGCGTCCACTATAAACAAAGCGGCCTTGCTGCAATGGCTTCGAAAGTACACCGACAACAACAACGAGAACGCCTTCAACAAGGCGATCGACGAGTTCACGATGAGCTGCGCCGGCTACTGCGTCGCCACCTACGTGCTGGGCATCGCCGACCGACATCCCGACAACATCATGGTCAAGAAGAGCGGGCAGCTCTTCCACATAGACTTCGGCCATTTCCTTGGACATTTCAAACAGAAGTACGGATTCAAACGGGAAAGGGTGCCCTTTGTGCTGACGCACGATTTTATATACGTCATCAATAAAGGTCAGAAGGGCTCCGGCGATAACAACGCTATCGACTTCAAAATATTCAAGGAGCATTGCGAAACTGCATTCAAAATCCTCCGCAAACATGGACATCTAATACTGTCGTTGTTCTCAATGATGATATCGACGGGGCTGCCGGAACTCAGCTCGGAGAAGGACTTGCAATACCTCAGAGATACATTAGTAATGGATCGGTCCGAAGAGAAAGCGATGGAGCATTTCCGTGAGAAGTTCTCGGACGCCCTCAAGAATTCGTGGAAGACATCCTTCAACTGGGTCTTCCATAACTGCGACAAGAACAACTGA
Protein
MVPAPSCPNTWDYWMSPQSDCVELTCLLPNSMYIPLRKSWDATFEDVKEDLWDRAVNYPLFGMLLEMNKYIFQFVNSLAVTEELDDERKRICDIKPVCGVLIIVKRSDGRPAEDCLNRDITHLIGKSLKVFDSLKNSEVNDYRMRMRYLAEESLLKRTHSTTLERLRYQCPLRLADQPEIPTTLKSHLNDQRFILVTKVMEITSYSSNVPATLTPQQHIEIVLRKLNNSMNLRSEHPHNYVLKVCGREEYLFGDYPLIQFLYIQEMLAVDGVPQVMTVSIDKLRLSLDREIPYMPERRRLTSEHSNTMRRKKNESSWNIQKNYTCRVQSACGLNVDPSRVVEVVCLGGIFHGGKPLCEVQKTKVAVISKEGVATWDEELTFPIKVHNLPRMARLCFGIYELSKIKSKWRGKDKEPVNRLTWANTMVFDYKEQLRTDGVRLAMWTHVADDTQGDDMVLHPLGTVVANPTPDSAVLQVMFSNYDCQYPILFPKQETVMAYSDHIESVSPDLMDRLCRDFERLRIIAEKDPMYEMHEQEKKDIWASRERFRVETPELLPKLLSCVEWGERTQASSVPRMLDEWPLLRVESALELLDYAYADAAVRSFAVKCLEKISDNDLMLYLLQLVQALKHESYLMCDLSVFLLQRAFKNMTIGHYLFWHLRSEMHVPSVSVRFRLMLEAYCRGSQEHIKILKQQITCLEKLKWVNQCVKEKKEISKARIALHQHLQEKHCVDALCEGFVSPLNPSFKCKRIKPEGCRVMDSKMRPLILNFENFDPTGNDIRIILKIGDDLRQDMFTLQMLRIMDRLWKNHGYDFRLNAYNCISIEYSVGMIEVVEDAETVANIQKQNAPFNAASTINKAALLQWLRKYTDNNNENAFNKAIDEFTMSCAGYCVATYVLGIADRHPDNIMVKKSGQLFHIDFGHFLGHFKQKYGFKRERVPFVLTHDFIYVINKGQKGSGDNNAIDFKIFKEHCETAFKILRKHGHLILSLFSMMISTGLPELSSEKDLQYLRDTLVMDRSEEKAMEHFREKFSDALKNSWKTSFNWVFHNCDKNN
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + ATP = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4,5-trisphosphate) + ADP + H(+)
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) + ATP = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol 3-phosphate) + ADP + H(+)
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) + ATP = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol 3-phosphate) + ADP + H(+)
Similarity
Belongs to the PI3/PI4-kinase family.
Uniprot
H9J086
A0A194Q421
A0A3S2LIU0
A0A194QSG1
A0A2H1W0S1
A0A212F0E0
+ More
A0A067RK76 A0A2J7PKL2 A0A1B6CVF3 A0A2A3ELR2 A0A087ZW41 A0A310SDB8 E0VFY9 A0A0C9R9T0 A0A151XF89 A0A0L7QVD9 A0A026WCF3 E2AJI7 A0A195FYH8 A0A158NHH3 E2C2K5 K7J7D6 F4WVN9 A0A154P0T2 A0A1Y1NB78 A0A0M9AAE9 A0A195D5M2 A0A195AV33 T1HZ23 A0A2H8TEQ4 A0A1W4WFE0 A0A0M5L080 A0A2S2PN65 E9JCJ6 J9JM92 D6X2N9 A0A195E2Y4 A0A1L8DMH7 A0A336MQM4 A0A336KXX9 U4U148 A0A1J1HI67 A0A146M2S1 A0A1S4F323 A0A182GI06 Q17GT9 N6U3K6 A0A1Q3F9V3 A0A0L0C2C7 E9HU25 A0A182JJK9 A0A1I8NE79 A0A1I8NRB0 B0WGF0 A0A1A9YFB3 U5EUF6 A0A1A9WY79 A0A336KYA5 Q7QEJ7 A0A0A1XHN1 W8BH04 A0A0N6VTH7 A0A0P4WTE3 A0A2S2N7D9 A0A182V5Y0 A0A1B6KS73 A0A346QRX4 A0A0P4WF28 A0A182HIU7 A0A0N6X4J6 A0A182XNN0 A0A182UFS2 D5KZJ2 T1ISF0 A0A0K8WEI3 A0A2M4A8R2 A0A034WN88 A0A182FK58 W5JCU6 A0A1L2BW25 B4K8F9 A0A087TVD8 B3M0A8 B4JUC0 B4LWM3 B4NFV1 B4II96 A0A3B0KV28 P91634 H1ZY91 B4GZD0 H1ZY88 Q29AY5 B4PPQ8 B3P349 A0A0M4F5A4 A0A0J7KCV9 B4QS08
A0A067RK76 A0A2J7PKL2 A0A1B6CVF3 A0A2A3ELR2 A0A087ZW41 A0A310SDB8 E0VFY9 A0A0C9R9T0 A0A151XF89 A0A0L7QVD9 A0A026WCF3 E2AJI7 A0A195FYH8 A0A158NHH3 E2C2K5 K7J7D6 F4WVN9 A0A154P0T2 A0A1Y1NB78 A0A0M9AAE9 A0A195D5M2 A0A195AV33 T1HZ23 A0A2H8TEQ4 A0A1W4WFE0 A0A0M5L080 A0A2S2PN65 E9JCJ6 J9JM92 D6X2N9 A0A195E2Y4 A0A1L8DMH7 A0A336MQM4 A0A336KXX9 U4U148 A0A1J1HI67 A0A146M2S1 A0A1S4F323 A0A182GI06 Q17GT9 N6U3K6 A0A1Q3F9V3 A0A0L0C2C7 E9HU25 A0A182JJK9 A0A1I8NE79 A0A1I8NRB0 B0WGF0 A0A1A9YFB3 U5EUF6 A0A1A9WY79 A0A336KYA5 Q7QEJ7 A0A0A1XHN1 W8BH04 A0A0N6VTH7 A0A0P4WTE3 A0A2S2N7D9 A0A182V5Y0 A0A1B6KS73 A0A346QRX4 A0A0P4WF28 A0A182HIU7 A0A0N6X4J6 A0A182XNN0 A0A182UFS2 D5KZJ2 T1ISF0 A0A0K8WEI3 A0A2M4A8R2 A0A034WN88 A0A182FK58 W5JCU6 A0A1L2BW25 B4K8F9 A0A087TVD8 B3M0A8 B4JUC0 B4LWM3 B4NFV1 B4II96 A0A3B0KV28 P91634 H1ZY91 B4GZD0 H1ZY88 Q29AY5 B4PPQ8 B3P349 A0A0M4F5A4 A0A0J7KCV9 B4QS08
EC Number
2.7.1.153
2.7.1.137
2.7.1.137
Pubmed
19121390
26354079
22118469
24845553
20566863
24508170
+ More
30249741 20798317 21347285 20075255 21719571 28004739 21282665 18362917 19820115 23537049 26823975 26483478 17510324 26108605 21292972 25315136 12364791 25830018 24495485 29544772 20132480 25348373 20920257 23761445 17994087 18057021 8978685 10731132 12537568 12537572 12537573 12537574 16110336 17043223 17569856 17569867 26109357 26109356 15632085 23185243 17550304
30249741 20798317 21347285 20075255 21719571 28004739 21282665 18362917 19820115 23537049 26823975 26483478 17510324 26108605 21292972 25315136 12364791 25830018 24495485 29544772 20132480 25348373 20920257 23761445 17994087 18057021 8978685 10731132 12537568 12537572 12537573 12537574 16110336 17043223 17569856 17569867 26109357 26109356 15632085 23185243 17550304
EMBL
BABH01019095
BABH01019096
KQ459472
KPJ00094.1
RSAL01000003
RVE54776.1
+ More
KQ461175 KPJ07920.1 ODYU01005640 SOQ46690.1 AGBW02011105 OWR47209.1 KK852575 KDR21011.1 NEVH01024536 PNF16872.1 GEDC01019779 JAS17519.1 KZ288215 PBC32434.1 KQ763441 OAD55097.1 AAZO01001977 DS235129 EEB12295.1 GBYB01009692 GBYB01011766 JAG79459.1 JAG81533.1 KQ982194 KYQ59052.1 KQ414727 KOC62607.1 KK107293 QOIP01000005 EZA53361.1 RLU22269.1 GL440027 EFN66382.1 KQ981208 KYN44889.1 ADTU01015824 GL452135 EFN77861.1 AAZX01005511 GL888393 EGI61709.1 KQ434792 KZC05432.1 GEZM01010700 JAV93895.1 KQ435713 KOX79243.1 KQ976870 KYN07739.1 KQ976737 KYM75917.1 ACPB03025045 ACPB03025046 GFXV01000782 MBW12587.1 KR075839 ALE20558.1 GGMR01018273 MBY30892.1 GL771844 EFZ09458.1 ABLF02031891 KQ971372 EFA09848.2 KQ979763 KYN19244.1 GFDF01006432 JAV07652.1 UFQS01001692 UFQT01001692 SSX11876.1 SSX31439.1 UFQS01001210 UFQT01001210 SSX09802.1 SSX29525.1 KB631865 ERL86797.1 CVRI01000004 CRK87595.1 GDHC01004541 JAQ14088.1 JXUM01011928 JXUM01011929 JXUM01011930 KQ560341 KXJ82960.1 CH477256 EAT45848.1 APGK01041002 KB740986 ENN76155.1 GFDL01010746 JAV24299.1 JRES01000988 KNC26382.1 GL732794 EFX64751.1 DS231926 EDS26947.1 GANO01002343 JAB57528.1 SSX09803.1 SSX29526.1 AAAB01008846 EAA06493.5 GBXI01014672 GBXI01008740 GBXI01008518 GBXI01008235 GBXI01003825 JAC99619.1 JAD05552.1 JAD05774.1 JAD06057.1 JAD10467.1 GAMC01014019 JAB92536.1 JX452100 AGK90307.1 GDRN01062052 JAI65119.1 GGMR01000456 MBY13075.1 GEBQ01025671 JAT14306.1 MH113349 AXR99085.1 GDRN01062053 JAI65118.1 APCN01004099 JX853773 AFU61123.1 GU724585 ADE44091.1 JH431430 GDHF01004639 GDHF01002802 JAI47675.1 JAI49512.1 GGFK01003872 MBW37193.1 GAKP01003150 JAC55802.1 ADMH02001755 ETN61168.1 KP843004 ALY05366.1 CH933806 EDW16541.1 KRG02183.1 KK116915 KFM69077.1 CH902617 EDV43114.1 KPU80091.1 CH916374 EDV91090.1 CH940650 EDW67688.1 KRF83413.1 KRF83414.1 CH964251 EDW83168.1 CH480842 EDW49640.1 OUUW01000013 SPP87818.1 AE014297 BT003173 DQ792599 DQ792600 Y09070 FR870118 FR870119 FR870121 FR870122 FR870124 FR870125 FR870126 FR870127 FR870128 AAF55792.1 AAN14359.1 AAO24928.1 ABG88073.1 ABG88075.1 AGB96150.1 CAA70291.1 CCB63076.1 CCB63077.1 CCB63079.1 CCB63080.1 CCB63082.1 CCB63083.1 CCB63084.1 CCB63085.1 CCB63086.1 FR870120 FR870123 CCB63078.1 CCB63081.1 CH479198 EDW28148.1 FR870117 CCB63075.1 CM000070 EAL27214.3 KRS99840.1 CM000160 EDW96158.1 KRK02941.1 CH954181 EDV48363.1 CP012526 ALC46695.1 LBMM01009539 KMQ88046.1 CM000364 EDX12204.1
KQ461175 KPJ07920.1 ODYU01005640 SOQ46690.1 AGBW02011105 OWR47209.1 KK852575 KDR21011.1 NEVH01024536 PNF16872.1 GEDC01019779 JAS17519.1 KZ288215 PBC32434.1 KQ763441 OAD55097.1 AAZO01001977 DS235129 EEB12295.1 GBYB01009692 GBYB01011766 JAG79459.1 JAG81533.1 KQ982194 KYQ59052.1 KQ414727 KOC62607.1 KK107293 QOIP01000005 EZA53361.1 RLU22269.1 GL440027 EFN66382.1 KQ981208 KYN44889.1 ADTU01015824 GL452135 EFN77861.1 AAZX01005511 GL888393 EGI61709.1 KQ434792 KZC05432.1 GEZM01010700 JAV93895.1 KQ435713 KOX79243.1 KQ976870 KYN07739.1 KQ976737 KYM75917.1 ACPB03025045 ACPB03025046 GFXV01000782 MBW12587.1 KR075839 ALE20558.1 GGMR01018273 MBY30892.1 GL771844 EFZ09458.1 ABLF02031891 KQ971372 EFA09848.2 KQ979763 KYN19244.1 GFDF01006432 JAV07652.1 UFQS01001692 UFQT01001692 SSX11876.1 SSX31439.1 UFQS01001210 UFQT01001210 SSX09802.1 SSX29525.1 KB631865 ERL86797.1 CVRI01000004 CRK87595.1 GDHC01004541 JAQ14088.1 JXUM01011928 JXUM01011929 JXUM01011930 KQ560341 KXJ82960.1 CH477256 EAT45848.1 APGK01041002 KB740986 ENN76155.1 GFDL01010746 JAV24299.1 JRES01000988 KNC26382.1 GL732794 EFX64751.1 DS231926 EDS26947.1 GANO01002343 JAB57528.1 SSX09803.1 SSX29526.1 AAAB01008846 EAA06493.5 GBXI01014672 GBXI01008740 GBXI01008518 GBXI01008235 GBXI01003825 JAC99619.1 JAD05552.1 JAD05774.1 JAD06057.1 JAD10467.1 GAMC01014019 JAB92536.1 JX452100 AGK90307.1 GDRN01062052 JAI65119.1 GGMR01000456 MBY13075.1 GEBQ01025671 JAT14306.1 MH113349 AXR99085.1 GDRN01062053 JAI65118.1 APCN01004099 JX853773 AFU61123.1 GU724585 ADE44091.1 JH431430 GDHF01004639 GDHF01002802 JAI47675.1 JAI49512.1 GGFK01003872 MBW37193.1 GAKP01003150 JAC55802.1 ADMH02001755 ETN61168.1 KP843004 ALY05366.1 CH933806 EDW16541.1 KRG02183.1 KK116915 KFM69077.1 CH902617 EDV43114.1 KPU80091.1 CH916374 EDV91090.1 CH940650 EDW67688.1 KRF83413.1 KRF83414.1 CH964251 EDW83168.1 CH480842 EDW49640.1 OUUW01000013 SPP87818.1 AE014297 BT003173 DQ792599 DQ792600 Y09070 FR870118 FR870119 FR870121 FR870122 FR870124 FR870125 FR870126 FR870127 FR870128 AAF55792.1 AAN14359.1 AAO24928.1 ABG88073.1 ABG88075.1 AGB96150.1 CAA70291.1 CCB63076.1 CCB63077.1 CCB63079.1 CCB63080.1 CCB63082.1 CCB63083.1 CCB63084.1 CCB63085.1 CCB63086.1 FR870120 FR870123 CCB63078.1 CCB63081.1 CH479198 EDW28148.1 FR870117 CCB63075.1 CM000070 EAL27214.3 KRS99840.1 CM000160 EDW96158.1 KRK02941.1 CH954181 EDV48363.1 CP012526 ALC46695.1 LBMM01009539 KMQ88046.1 CM000364 EDX12204.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000007151
UP000027135
+ More
UP000235965 UP000242457 UP000005203 UP000009046 UP000075809 UP000053825 UP000053097 UP000279307 UP000000311 UP000078541 UP000005205 UP000008237 UP000002358 UP000007755 UP000076502 UP000053105 UP000078542 UP000078540 UP000015103 UP000192223 UP000007819 UP000007266 UP000078492 UP000030742 UP000183832 UP000069940 UP000249989 UP000008820 UP000019118 UP000037069 UP000000305 UP000075880 UP000095301 UP000095300 UP000002320 UP000092443 UP000091820 UP000007062 UP000075903 UP000075840 UP000076407 UP000075902 UP000069272 UP000000673 UP000009192 UP000054359 UP000007801 UP000001070 UP000008792 UP000007798 UP000001292 UP000268350 UP000000803 UP000008744 UP000001819 UP000002282 UP000008711 UP000092553 UP000036403 UP000000304
UP000235965 UP000242457 UP000005203 UP000009046 UP000075809 UP000053825 UP000053097 UP000279307 UP000000311 UP000078541 UP000005205 UP000008237 UP000002358 UP000007755 UP000076502 UP000053105 UP000078542 UP000078540 UP000015103 UP000192223 UP000007819 UP000007266 UP000078492 UP000030742 UP000183832 UP000069940 UP000249989 UP000008820 UP000019118 UP000037069 UP000000305 UP000075880 UP000095301 UP000095300 UP000002320 UP000092443 UP000091820 UP000007062 UP000075903 UP000075840 UP000076407 UP000075902 UP000069272 UP000000673 UP000009192 UP000054359 UP000007801 UP000001070 UP000008792 UP000007798 UP000001292 UP000268350 UP000000803 UP000008744 UP000001819 UP000002282 UP000008711 UP000092553 UP000036403 UP000000304
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR015433 PI_Kinase
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR003113 PI3K_adapt-bd_dom
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR029071 Ubiquitin-like_domsf
IPR008290 PI3K_Vps34
IPR036940 PI3/4_kinase_cat_sf
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR005284 Pigment_permease/Abcg
IPR013525 ABC_2_trans
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR015433 PI_Kinase
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR003113 PI3K_adapt-bd_dom
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR029071 Ubiquitin-like_domsf
IPR008290 PI3K_Vps34
IPR036940 PI3/4_kinase_cat_sf
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR005284 Pigment_permease/Abcg
IPR013525 ABC_2_trans
IPR005828 MFS_sugar_transport-like
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J086
A0A194Q421
A0A3S2LIU0
A0A194QSG1
A0A2H1W0S1
A0A212F0E0
+ More
A0A067RK76 A0A2J7PKL2 A0A1B6CVF3 A0A2A3ELR2 A0A087ZW41 A0A310SDB8 E0VFY9 A0A0C9R9T0 A0A151XF89 A0A0L7QVD9 A0A026WCF3 E2AJI7 A0A195FYH8 A0A158NHH3 E2C2K5 K7J7D6 F4WVN9 A0A154P0T2 A0A1Y1NB78 A0A0M9AAE9 A0A195D5M2 A0A195AV33 T1HZ23 A0A2H8TEQ4 A0A1W4WFE0 A0A0M5L080 A0A2S2PN65 E9JCJ6 J9JM92 D6X2N9 A0A195E2Y4 A0A1L8DMH7 A0A336MQM4 A0A336KXX9 U4U148 A0A1J1HI67 A0A146M2S1 A0A1S4F323 A0A182GI06 Q17GT9 N6U3K6 A0A1Q3F9V3 A0A0L0C2C7 E9HU25 A0A182JJK9 A0A1I8NE79 A0A1I8NRB0 B0WGF0 A0A1A9YFB3 U5EUF6 A0A1A9WY79 A0A336KYA5 Q7QEJ7 A0A0A1XHN1 W8BH04 A0A0N6VTH7 A0A0P4WTE3 A0A2S2N7D9 A0A182V5Y0 A0A1B6KS73 A0A346QRX4 A0A0P4WF28 A0A182HIU7 A0A0N6X4J6 A0A182XNN0 A0A182UFS2 D5KZJ2 T1ISF0 A0A0K8WEI3 A0A2M4A8R2 A0A034WN88 A0A182FK58 W5JCU6 A0A1L2BW25 B4K8F9 A0A087TVD8 B3M0A8 B4JUC0 B4LWM3 B4NFV1 B4II96 A0A3B0KV28 P91634 H1ZY91 B4GZD0 H1ZY88 Q29AY5 B4PPQ8 B3P349 A0A0M4F5A4 A0A0J7KCV9 B4QS08
A0A067RK76 A0A2J7PKL2 A0A1B6CVF3 A0A2A3ELR2 A0A087ZW41 A0A310SDB8 E0VFY9 A0A0C9R9T0 A0A151XF89 A0A0L7QVD9 A0A026WCF3 E2AJI7 A0A195FYH8 A0A158NHH3 E2C2K5 K7J7D6 F4WVN9 A0A154P0T2 A0A1Y1NB78 A0A0M9AAE9 A0A195D5M2 A0A195AV33 T1HZ23 A0A2H8TEQ4 A0A1W4WFE0 A0A0M5L080 A0A2S2PN65 E9JCJ6 J9JM92 D6X2N9 A0A195E2Y4 A0A1L8DMH7 A0A336MQM4 A0A336KXX9 U4U148 A0A1J1HI67 A0A146M2S1 A0A1S4F323 A0A182GI06 Q17GT9 N6U3K6 A0A1Q3F9V3 A0A0L0C2C7 E9HU25 A0A182JJK9 A0A1I8NE79 A0A1I8NRB0 B0WGF0 A0A1A9YFB3 U5EUF6 A0A1A9WY79 A0A336KYA5 Q7QEJ7 A0A0A1XHN1 W8BH04 A0A0N6VTH7 A0A0P4WTE3 A0A2S2N7D9 A0A182V5Y0 A0A1B6KS73 A0A346QRX4 A0A0P4WF28 A0A182HIU7 A0A0N6X4J6 A0A182XNN0 A0A182UFS2 D5KZJ2 T1ISF0 A0A0K8WEI3 A0A2M4A8R2 A0A034WN88 A0A182FK58 W5JCU6 A0A1L2BW25 B4K8F9 A0A087TVD8 B3M0A8 B4JUC0 B4LWM3 B4NFV1 B4II96 A0A3B0KV28 P91634 H1ZY91 B4GZD0 H1ZY88 Q29AY5 B4PPQ8 B3P349 A0A0M4F5A4 A0A0J7KCV9 B4QS08
PDB
5IS5
E-value=0,
Score=1876
Ontologies
PATHWAY
00562
Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04070 Phosphatidylinositol signaling system - Bombyx mori (domestic silkworm)
04140 Autophagy - animal - Bombyx mori (domestic silkworm)
04150 mTOR signaling pathway - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
04933 AGE-RAGE signaling pathway in diabetic complications - Bombyx mori (domestic silkworm)
GO
GO:0052812
GO:0005524
GO:0016303
GO:0046934
GO:0048015
GO:0005942
GO:0005886
GO:0016477
GO:0005737
GO:0036092
GO:0035005
GO:0016020
GO:0046854
GO:0014065
GO:0016301
GO:0016021
GO:0055085
GO:0016887
GO:0022857
GO:0055115
GO:0007390
GO:0009267
GO:0035264
GO:1904801
GO:0048169
GO:0035212
GO:0036335
GO:0030707
GO:0051823
GO:0008286
GO:0060074
GO:0006979
GO:0007552
GO:0055088
GO:0030307
GO:0005975
GO:0071902
GO:0010884
GO:0048010
GO:0006914
GO:0042593
GO:0040014
GO:0051897
GO:0005943
GO:0030536
GO:0045572
GO:0007436
GO:0048813
GO:0007346
GO:0010507
GO:0045727
GO:2000377
GO:2000252
GO:0045793
GO:0042127
GO:0045887
GO:0036099
GO:0050773
GO:0035099
GO:1904263
GO:0008361
GO:0006629
GO:0002164
GO:0048167
GO:0046622
GO:0048477
GO:0002168
GO:0005829
GO:0035004
GO:0005515
GO:0005488
GO:0016765
GO:0005634
GO:0006355
GO:0016818
GO:0003676
GO:0008026
GO:0016597
PANTHER
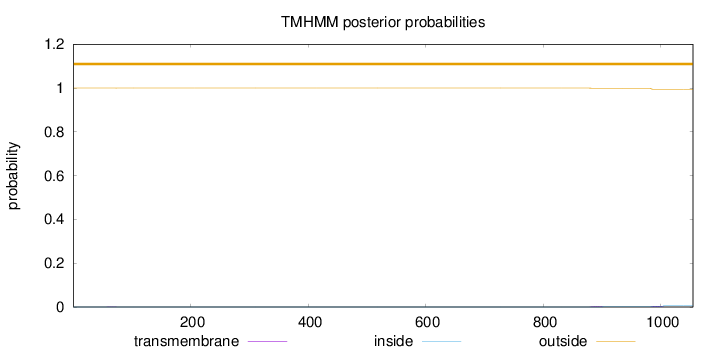
Topology
Length:
1055
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11699
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.00018
outside
1 - 1055
Population Genetic Test Statistics
Pi
173.915935
Theta
158.683303
Tajima's D
0.499049
CLR
1.244445
CSRT
0.512274386280686
Interpretation
Uncertain
