Gene
KWMTBOMO05906
Pre Gene Modal
BGIBMGA002922
Annotation
ATP_dependent_transmembrane_transporter_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.592
Sequence
CDS
ATGATTGGGGTATTCTGCTCTGAACTACCAATCTTCATCAGAGAGCACCACTCGGGAATGTATCGAGCCGACGTCTATTTCTTATCGAAGACGTTAGCCGAAGCTCCCGTTTTCGCCACTATACCATTAGTCTTCACAACTATAGCTTATTACATGATCGGATTGAACCCAGATCCGAAGAGGTTCTTCATCGCTTCAGGATTGGCTGCTCTGGTTACGAACGTCGCTACTTCGTTCGGTTATTTAATATCCTGTGCCAGCAGCAGCGTCAGTATGGCAGCCTCCGTTGGTCCTCCAATCATCATCCCCTTTATGCTATTTGGTGGATTCTTCTTGAATTCTGGGTGA
Protein
MIGVFCSELPIFIREHHSGMYRADVYFLSKTLAEAPVFATIPLVFTTIAYYMIGLNPDPKRFFIASGLAALVTNVATSFGYLISCASSSVSMAASVGPPIIIPFMLFGGFFLNSG
Summary
Similarity
Belongs to the PI3/PI4-kinase family.
Uniprot
A0A2A4J524
A0A222AJ59
I4DQW9
Q9NH94
B7XCV5
H9J087
+ More
A0A2A4IX22 B3GQX5 A0A1C8V5Z8 A0A2H1VXR6 A0A1C8V600 A0A3S2NRU7 A0A194Q3I4 A0A194QRF1 A0A0C5PTD7 A0A212F0E3 A0A1B6K9V1 A0A195E2Y4 A0A195D4B1 A0A026WBC3 A0A1B6GUX8 A0A3L8DP96 A0A2J7QE54 A0A2Z5ZBX4 A0A158NHH2 A0A067RAU0 A0A195FWS3 F4WVN8 A0A1B6ICC2 A0A151XF99 A0A195AUR0 A0A0J7KSS0 A0A0M9A9I2 A0A0L7QVP7 A0A154P273 A0A232FDM6 A0A0C9PHK2 A0A2A3EMK8 A0A0C9QIB5 A0A1B0YWI7 A0A2S0X9Z2 T1I8T7 A0A1B6CC40 E2AJI8 J9K5S2 Q60FC5 A0A2S2QA04 A0A3R5WTC8 A0A2H8TYR6 A0A0P5KR80 A0A0P6AKJ6 A0A0P5J785 A0A158V6Z1 A0A0A9XU47 A0A0P6H2N5 A0A3Q0IQC3 A0A0P6CF28 B4GYI0 B4GYI4 A0A0N8ADT7 A0A0P5PD96 A0A3B0KIY5 A0A146M420 B5DLT3 A0A3B0KPD8 A0A0R3NZL1 A0A0N8E341 A0A0P5ISF6 A0A0P5H647 A0A0K8SNK1 A0A0P5M8S0 A0A0P5UDH5 A0A0P4Y5T3 A0A1I8MED8 A0A1D2NLM5 T1JB20 A0A0P5FQW1 A0A0N8AR28 A0A0P5RD50 A0A0P5FYW1 A0A0P5KWN4 A0A0N8C079 A0A0P5R3F0 A0A0P5LL12 O77423 A0A034VCM1 A0A0P5LS18 A0A0K8W6U9 A0A0N8B7D9 A0A0N8B004 A0A0P5HWS2 A0A0P5JC00 A0A0Q9W770 B4MG61 Q8IS30 B4JJE3 A0A0P6IIG0 A0A0P5JMP6 A0A0P5MQS4 A0A0P5PXA3
A0A2A4IX22 B3GQX5 A0A1C8V5Z8 A0A2H1VXR6 A0A1C8V600 A0A3S2NRU7 A0A194Q3I4 A0A194QRF1 A0A0C5PTD7 A0A212F0E3 A0A1B6K9V1 A0A195E2Y4 A0A195D4B1 A0A026WBC3 A0A1B6GUX8 A0A3L8DP96 A0A2J7QE54 A0A2Z5ZBX4 A0A158NHH2 A0A067RAU0 A0A195FWS3 F4WVN8 A0A1B6ICC2 A0A151XF99 A0A195AUR0 A0A0J7KSS0 A0A0M9A9I2 A0A0L7QVP7 A0A154P273 A0A232FDM6 A0A0C9PHK2 A0A2A3EMK8 A0A0C9QIB5 A0A1B0YWI7 A0A2S0X9Z2 T1I8T7 A0A1B6CC40 E2AJI8 J9K5S2 Q60FC5 A0A2S2QA04 A0A3R5WTC8 A0A2H8TYR6 A0A0P5KR80 A0A0P6AKJ6 A0A0P5J785 A0A158V6Z1 A0A0A9XU47 A0A0P6H2N5 A0A3Q0IQC3 A0A0P6CF28 B4GYI0 B4GYI4 A0A0N8ADT7 A0A0P5PD96 A0A3B0KIY5 A0A146M420 B5DLT3 A0A3B0KPD8 A0A0R3NZL1 A0A0N8E341 A0A0P5ISF6 A0A0P5H647 A0A0K8SNK1 A0A0P5M8S0 A0A0P5UDH5 A0A0P4Y5T3 A0A1I8MED8 A0A1D2NLM5 T1JB20 A0A0P5FQW1 A0A0N8AR28 A0A0P5RD50 A0A0P5FYW1 A0A0P5KWN4 A0A0N8C079 A0A0P5R3F0 A0A0P5LL12 O77423 A0A034VCM1 A0A0P5LS18 A0A0K8W6U9 A0A0N8B7D9 A0A0N8B004 A0A0P5HWS2 A0A0P5JC00 A0A0Q9W770 B4MG61 Q8IS30 B4JJE3 A0A0P6IIG0 A0A0P5JMP6 A0A0P5MQS4 A0A0P5PXA3
Pubmed
EMBL
NWSH01003289
PCG66614.1
KY564059
ASO76408.1
AK404815
BAM20309.1
+ More
AF229609 AAF61569.1 AB445460 BAH03523.1 BABH01019098 BABH01019099 BABH01019100 BABH01019101 BABH01019102 BABH01019103 BABH01019104 BABH01019105 NWSH01005714 PCG64016.1 EU722903 ACD99701.1 KU754477 ANW09739.1 ODYU01005100 SOQ45637.1 KU754476 ANW09738.1 RSAL01000003 RVE54777.1 KQ459472 KPJ00092.1 KQ461175 KPJ07919.1 KM667971 AJQ21779.1 AGBW02011105 OWR47210.1 GEBQ01031766 JAT08211.1 KQ979763 KYN19244.1 KQ976870 KYN07740.1 KK107293 EZA53360.1 GECZ01003552 JAS66217.1 QOIP01000005 RLU22270.1 NEVH01015327 PNF26870.1 LC368084 BBC77943.1 ADTU01015824 KK852575 KDR20987.1 KQ981208 KYN44888.1 GL888393 EGI61708.1 GECU01023128 JAS84578.1 KQ982194 KYQ59053.1 KQ976737 KYM75916.1 LBMM01003652 KMQ93279.1 KQ435713 KOX79246.1 KQ414727 KOC62606.1 KQ434792 KZC05434.1 NNAY01000361 OXU28894.1 GBYB01000323 JAG70090.1 KZ288215 PBC32436.1 GBYB01000322 JAG70089.1 KU376474 ANO53447.1 MH105806 AWC08636.1 ACPB03022306 GEDC01026272 JAS11026.1 GL440027 EFN66383.1 ABLF02031566 AB191788 BAD54748.1 GGMS01005364 MBY74567.1 MH172526 QAA95933.1 GFXV01007225 MBW19030.1 GDIQ01180846 JAK70879.1 GDIP01041545 JAM62170.1 GDIQ01205549 GDIQ01205548 JAK46176.1 KF828804 AIN44121.1 GBHO01021188 GBHO01021186 GBHO01021185 JAG22416.1 JAG22418.1 JAG22419.1 GDIQ01026153 JAN68584.1 GDIP01016265 JAM87450.1 CH479197 EDW27836.1 EDW27840.1 GDIP01157241 JAJ66161.1 GDIQ01130277 JAL21449.1 OUUW01000015 SPP88490.1 GDHC01004115 JAQ14514.1 CH379064 EDY72595.1 SPP88489.1 KRT06475.1 GDIQ01066540 JAN28197.1 GDIQ01217487 GDIQ01199753 JAK34238.1 GDIQ01232276 JAK19449.1 GBRD01011135 GBRD01007802 JAG54689.1 GDIQ01161752 JAK89973.1 GDIP01115976 JAL87738.1 GDIP01232080 JAI91321.1 LJIJ01000010 ODN06152.1 JH432008 GDIQ01251237 GDIQ01251236 JAK00488.1 GDIQ01251235 JAK00490.1 GDIQ01121918 JAL29808.1 GDIQ01252056 GDIQ01252055 JAJ99669.1 GDIQ01203902 GDIQ01203901 JAK47823.1 GDIQ01127627 GDIQ01126033 JAL24099.1 GDIQ01126032 JAL25694.1 GDIQ01168113 JAK83612.1 U97104 AAC61893.1 GAKP01017893 GAKP01017891 JAC41061.1 GDIQ01168114 JAK83611.1 GDHF01033402 GDHF01005555 JAI18912.1 JAI46759.1 GDIQ01205547 JAK46178.1 GDIQ01226227 JAK25498.1 GDIQ01223877 JAK27848.1 GDIQ01202296 GDIQ01202295 GDIQ01202294 JAK49429.1 CH940672 KRF77745.1 EDW57384.1 AH012033 GBXI01016111 AAN38825.1 JAC98180.1 CH916370 EDV99695.1 GDIQ01030805 JAN63932.1 GDIQ01201000 GDIQ01200999 GDIQ01200998 GDIQ01199754 GDIQ01175450 GDIQ01098684 GDIQ01059162 JAK50726.1 GDIQ01268145 GDIQ01233735 GDIQ01155472 GDIQ01152717 GDIQ01106933 JAK99008.1 GDIQ01142919 GDIP01069799 JAL08807.1 JAM33916.1
AF229609 AAF61569.1 AB445460 BAH03523.1 BABH01019098 BABH01019099 BABH01019100 BABH01019101 BABH01019102 BABH01019103 BABH01019104 BABH01019105 NWSH01005714 PCG64016.1 EU722903 ACD99701.1 KU754477 ANW09739.1 ODYU01005100 SOQ45637.1 KU754476 ANW09738.1 RSAL01000003 RVE54777.1 KQ459472 KPJ00092.1 KQ461175 KPJ07919.1 KM667971 AJQ21779.1 AGBW02011105 OWR47210.1 GEBQ01031766 JAT08211.1 KQ979763 KYN19244.1 KQ976870 KYN07740.1 KK107293 EZA53360.1 GECZ01003552 JAS66217.1 QOIP01000005 RLU22270.1 NEVH01015327 PNF26870.1 LC368084 BBC77943.1 ADTU01015824 KK852575 KDR20987.1 KQ981208 KYN44888.1 GL888393 EGI61708.1 GECU01023128 JAS84578.1 KQ982194 KYQ59053.1 KQ976737 KYM75916.1 LBMM01003652 KMQ93279.1 KQ435713 KOX79246.1 KQ414727 KOC62606.1 KQ434792 KZC05434.1 NNAY01000361 OXU28894.1 GBYB01000323 JAG70090.1 KZ288215 PBC32436.1 GBYB01000322 JAG70089.1 KU376474 ANO53447.1 MH105806 AWC08636.1 ACPB03022306 GEDC01026272 JAS11026.1 GL440027 EFN66383.1 ABLF02031566 AB191788 BAD54748.1 GGMS01005364 MBY74567.1 MH172526 QAA95933.1 GFXV01007225 MBW19030.1 GDIQ01180846 JAK70879.1 GDIP01041545 JAM62170.1 GDIQ01205549 GDIQ01205548 JAK46176.1 KF828804 AIN44121.1 GBHO01021188 GBHO01021186 GBHO01021185 JAG22416.1 JAG22418.1 JAG22419.1 GDIQ01026153 JAN68584.1 GDIP01016265 JAM87450.1 CH479197 EDW27836.1 EDW27840.1 GDIP01157241 JAJ66161.1 GDIQ01130277 JAL21449.1 OUUW01000015 SPP88490.1 GDHC01004115 JAQ14514.1 CH379064 EDY72595.1 SPP88489.1 KRT06475.1 GDIQ01066540 JAN28197.1 GDIQ01217487 GDIQ01199753 JAK34238.1 GDIQ01232276 JAK19449.1 GBRD01011135 GBRD01007802 JAG54689.1 GDIQ01161752 JAK89973.1 GDIP01115976 JAL87738.1 GDIP01232080 JAI91321.1 LJIJ01000010 ODN06152.1 JH432008 GDIQ01251237 GDIQ01251236 JAK00488.1 GDIQ01251235 JAK00490.1 GDIQ01121918 JAL29808.1 GDIQ01252056 GDIQ01252055 JAJ99669.1 GDIQ01203902 GDIQ01203901 JAK47823.1 GDIQ01127627 GDIQ01126033 JAL24099.1 GDIQ01126032 JAL25694.1 GDIQ01168113 JAK83612.1 U97104 AAC61893.1 GAKP01017893 GAKP01017891 JAC41061.1 GDIQ01168114 JAK83611.1 GDHF01033402 GDHF01005555 JAI18912.1 JAI46759.1 GDIQ01205547 JAK46178.1 GDIQ01226227 JAK25498.1 GDIQ01223877 JAK27848.1 GDIQ01202296 GDIQ01202295 GDIQ01202294 JAK49429.1 CH940672 KRF77745.1 EDW57384.1 AH012033 GBXI01016111 AAN38825.1 JAC98180.1 CH916370 EDV99695.1 GDIQ01030805 JAN63932.1 GDIQ01201000 GDIQ01200999 GDIQ01200998 GDIQ01199754 GDIQ01175450 GDIQ01098684 GDIQ01059162 JAK50726.1 GDIQ01268145 GDIQ01233735 GDIQ01155472 GDIQ01152717 GDIQ01106933 JAK99008.1 GDIQ01142919 GDIP01069799 JAL08807.1 JAM33916.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000078492 UP000078542 UP000053097 UP000279307 UP000235965 UP000005205 UP000027135 UP000078541 UP000007755 UP000075809 UP000078540 UP000036403 UP000053105 UP000053825 UP000076502 UP000215335 UP000242457 UP000015103 UP000000311 UP000007819 UP000079169 UP000008744 UP000268350 UP000001819 UP000095301 UP000094527 UP000008792 UP000001070
UP000078492 UP000078542 UP000053097 UP000279307 UP000235965 UP000005205 UP000027135 UP000078541 UP000007755 UP000075809 UP000078540 UP000036403 UP000053105 UP000053825 UP000076502 UP000215335 UP000242457 UP000015103 UP000000311 UP000007819 UP000079169 UP000008744 UP000268350 UP000001819 UP000095301 UP000094527 UP000008792 UP000001070
Pfam
Interpro
IPR013525
ABC_2_trans
+ More
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR005284 Pigment_permease/Abcg
IPR003593 AAA+_ATPase
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR003113 PI3K_adapt-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR025110 AMP-bd_C
IPR001736 PLipase_D/transphosphatidylase
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR005284 Pigment_permease/Abcg
IPR003593 AAA+_ATPase
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR003113 PI3K_adapt-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR025110 AMP-bd_C
IPR001736 PLipase_D/transphosphatidylase
Gene 3D
ProteinModelPortal
A0A2A4J524
A0A222AJ59
I4DQW9
Q9NH94
B7XCV5
H9J087
+ More
A0A2A4IX22 B3GQX5 A0A1C8V5Z8 A0A2H1VXR6 A0A1C8V600 A0A3S2NRU7 A0A194Q3I4 A0A194QRF1 A0A0C5PTD7 A0A212F0E3 A0A1B6K9V1 A0A195E2Y4 A0A195D4B1 A0A026WBC3 A0A1B6GUX8 A0A3L8DP96 A0A2J7QE54 A0A2Z5ZBX4 A0A158NHH2 A0A067RAU0 A0A195FWS3 F4WVN8 A0A1B6ICC2 A0A151XF99 A0A195AUR0 A0A0J7KSS0 A0A0M9A9I2 A0A0L7QVP7 A0A154P273 A0A232FDM6 A0A0C9PHK2 A0A2A3EMK8 A0A0C9QIB5 A0A1B0YWI7 A0A2S0X9Z2 T1I8T7 A0A1B6CC40 E2AJI8 J9K5S2 Q60FC5 A0A2S2QA04 A0A3R5WTC8 A0A2H8TYR6 A0A0P5KR80 A0A0P6AKJ6 A0A0P5J785 A0A158V6Z1 A0A0A9XU47 A0A0P6H2N5 A0A3Q0IQC3 A0A0P6CF28 B4GYI0 B4GYI4 A0A0N8ADT7 A0A0P5PD96 A0A3B0KIY5 A0A146M420 B5DLT3 A0A3B0KPD8 A0A0R3NZL1 A0A0N8E341 A0A0P5ISF6 A0A0P5H647 A0A0K8SNK1 A0A0P5M8S0 A0A0P5UDH5 A0A0P4Y5T3 A0A1I8MED8 A0A1D2NLM5 T1JB20 A0A0P5FQW1 A0A0N8AR28 A0A0P5RD50 A0A0P5FYW1 A0A0P5KWN4 A0A0N8C079 A0A0P5R3F0 A0A0P5LL12 O77423 A0A034VCM1 A0A0P5LS18 A0A0K8W6U9 A0A0N8B7D9 A0A0N8B004 A0A0P5HWS2 A0A0P5JC00 A0A0Q9W770 B4MG61 Q8IS30 B4JJE3 A0A0P6IIG0 A0A0P5JMP6 A0A0P5MQS4 A0A0P5PXA3
A0A2A4IX22 B3GQX5 A0A1C8V5Z8 A0A2H1VXR6 A0A1C8V600 A0A3S2NRU7 A0A194Q3I4 A0A194QRF1 A0A0C5PTD7 A0A212F0E3 A0A1B6K9V1 A0A195E2Y4 A0A195D4B1 A0A026WBC3 A0A1B6GUX8 A0A3L8DP96 A0A2J7QE54 A0A2Z5ZBX4 A0A158NHH2 A0A067RAU0 A0A195FWS3 F4WVN8 A0A1B6ICC2 A0A151XF99 A0A195AUR0 A0A0J7KSS0 A0A0M9A9I2 A0A0L7QVP7 A0A154P273 A0A232FDM6 A0A0C9PHK2 A0A2A3EMK8 A0A0C9QIB5 A0A1B0YWI7 A0A2S0X9Z2 T1I8T7 A0A1B6CC40 E2AJI8 J9K5S2 Q60FC5 A0A2S2QA04 A0A3R5WTC8 A0A2H8TYR6 A0A0P5KR80 A0A0P6AKJ6 A0A0P5J785 A0A158V6Z1 A0A0A9XU47 A0A0P6H2N5 A0A3Q0IQC3 A0A0P6CF28 B4GYI0 B4GYI4 A0A0N8ADT7 A0A0P5PD96 A0A3B0KIY5 A0A146M420 B5DLT3 A0A3B0KPD8 A0A0R3NZL1 A0A0N8E341 A0A0P5ISF6 A0A0P5H647 A0A0K8SNK1 A0A0P5M8S0 A0A0P5UDH5 A0A0P4Y5T3 A0A1I8MED8 A0A1D2NLM5 T1JB20 A0A0P5FQW1 A0A0N8AR28 A0A0P5RD50 A0A0P5FYW1 A0A0P5KWN4 A0A0N8C079 A0A0P5R3F0 A0A0P5LL12 O77423 A0A034VCM1 A0A0P5LS18 A0A0K8W6U9 A0A0N8B7D9 A0A0N8B004 A0A0P5HWS2 A0A0P5JC00 A0A0Q9W770 B4MG61 Q8IS30 B4JJE3 A0A0P6IIG0 A0A0P5JMP6 A0A0P5MQS4 A0A0P5PXA3
PDB
6HCO
E-value=5.20712e-08,
Score=130
Ontologies
GO
PANTHER
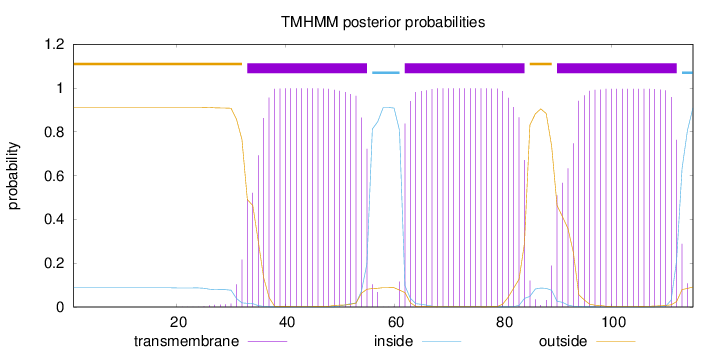
Topology
Length:
115
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.54689
Exp number, first 60 AAs:
21.54261
Total prob of N-in:
0.08743
POSSIBLE N-term signal
sequence
outside
1 - 32
TMhelix
33 - 55
inside
56 - 61
TMhelix
62 - 84
outside
85 - 89
TMhelix
90 - 112
inside
113 - 115
Population Genetic Test Statistics
Pi
270.553673
Theta
19.762943
Tajima's D
0.588026
CLR
0.64577
CSRT
0.545422728863557
Interpretation
Uncertain
