Gene
KWMTBOMO05904
Pre Gene Modal
BGIBMGA002922
Annotation
ATP_dependent_transmembrane_transporter_protein_[Bombyx_mori]
Full name
Protein white
Location in the cell
PlasmaMembrane Reliability : 3.933
Sequence
CDS
ATGAGAGAGAAACGCGCGATCAGTACGAGGCTTTGTGGCGGACTGCCGGCGCAGCAGCCCGGCGTCACCTGCGATAACGCGGCCGCGCGTTGGCTCCTGATTGGCGCGCGCACGCATCACCTGATTCTTGCGCGCAGGTCCGTCCCACCGTACTTAAGTTGGATATCGTATCTATCCTGGTTCCACTACGGCAACGAAGCGCTTCTCATCAACCAGTGGGCCGGCGTGGAGACTATCGCCTGCACAAGGGAGAACTTCACCTGTCCAGCTTCGGGACAGGTCGTACTGGAGACGCTTAGCTTTTCACAGGACGATTTCGCGATGGACGTGGTGAACATGATCTTGTTATTCGTCGGCTTCAGATTTCTGGCGTACCTCGCCCTGCTGTGGAGGACGCGCAGGGCCAAGTGA
Protein
MREKRAISTRLCGGLPAQQPGVTCDNAAARWLLIGARTHHLILARRSVPPYLSWISYLSWFHYGNEALLINQWAGVETIACTRENFTCPASGQVVLETLSFSQDDFAMDVVNMILLFVGFRFLAYLALLWRTRRAK
Summary
Description
Part of a membrane-spanning permease system necessary for the transport of pigment precursors into pigment cells responsible for eye color. White dimerize with brown for the transport of guanine. Scarlet and white complex transports a metabolic intermediate (such as 3-hydroxy kynurenine) from the cytoplasm into the pigment granules.
Subunit
Heterodimer of white with either brown or scarlet.
Similarity
Belongs to the PI3/PI4-kinase family.
Belongs to the ABC transporter superfamily. ABCG family. Eye pigment precursor importer (TC 3.A.1.204) subfamily.
Belongs to the ABC transporter superfamily. ABCG family. Eye pigment precursor importer (TC 3.A.1.204) subfamily.
Keywords
ATP-binding
Complete proteome
Membrane
Nucleotide-binding
Pigment
Reference proteome
Transmembrane
Transmembrane helix
Transport
Glycoprotein
Feature
chain Protein white
Uniprot
Q9NH94
H9J087
B7XCV5
B3GQX5
A0A212F0E3
A0A0C5PTD7
+ More
A0A194Q3I4 A0A194QRF1 A0A3R5WTC8 T1GGV8 A0A026WBC3 A0A3L8DP96 F4WVN8 A0A195D4B1 A0A151XF99 B4GYI5 A0A195FWS3 A0A158NHH2 A0A195AUR0 A0A2Z5ZBX4 A0A195E2Y4 A0A0L7QVP7 A0A0C9PHK2 A0A1B0CB12 A0A0C9QIB5 A0A0Q9W770 B4MG61 A0A0M4ETI1 A0A2S0X9Z2 A0A232FDM6 B4R3U7 Q60FC5 A0A158V6Z1 A0A1W4VF76 A0A1B0YWI7 B4L879 A0A3B0KIY5 A0A3B0KPD8 A0A0M9A9I2 B5DLT3 A0A0R3NZL1 P10090 B3NTX4 B3MWF6 A0A0R1EA29 A8E1U1 B4Q0V6 Q2LD54 A0A346A6M2 Q94960 Q8IS30 B4JJE3 W8AX98 A0A034VCM1 Q17320 A0A0K8VYU6 O77423 A0A0K8W6U9 B4NDV8 A0A1I8P5B8 Q8WRR1 Q8WRF2 A0A067RAU0 Q9BH97 E2C2K3 A0A1I8MED8 T1PA82 A0A087ZW42 A0A1A9W968 A0A1B0G2T2 A0A1A9XVI0 A0A1B0B3W9 A0A1A9V0K8 A0A1B0A8G3 A0A0J7KSS0
A0A194Q3I4 A0A194QRF1 A0A3R5WTC8 T1GGV8 A0A026WBC3 A0A3L8DP96 F4WVN8 A0A195D4B1 A0A151XF99 B4GYI5 A0A195FWS3 A0A158NHH2 A0A195AUR0 A0A2Z5ZBX4 A0A195E2Y4 A0A0L7QVP7 A0A0C9PHK2 A0A1B0CB12 A0A0C9QIB5 A0A0Q9W770 B4MG61 A0A0M4ETI1 A0A2S0X9Z2 A0A232FDM6 B4R3U7 Q60FC5 A0A158V6Z1 A0A1W4VF76 A0A1B0YWI7 B4L879 A0A3B0KIY5 A0A3B0KPD8 A0A0M9A9I2 B5DLT3 A0A0R3NZL1 P10090 B3NTX4 B3MWF6 A0A0R1EA29 A8E1U1 B4Q0V6 Q2LD54 A0A346A6M2 Q94960 Q8IS30 B4JJE3 W8AX98 A0A034VCM1 Q17320 A0A0K8VYU6 O77423 A0A0K8W6U9 B4NDV8 A0A1I8P5B8 Q8WRR1 Q8WRF2 A0A067RAU0 Q9BH97 E2C2K3 A0A1I8MED8 T1PA82 A0A087ZW42 A0A1A9W968 A0A1B0G2T2 A0A1A9XVI0 A0A1B0B3W9 A0A1A9V0K8 A0A1B0A8G3 A0A0J7KSS0
Pubmed
11016828
19121390
18996197
22118469
25636859
26354079
+ More
24508170 30249741 21719571 17994087 21347285 29654886 28648823 15705502 15632085 2109311 6084717 11156992 10731132 12537572 10731137 2503416 11294610 17550304 15972463 29187976 8601488 8889526 25830018 24495485 25348373 8533095 9359576 18362917 19820115 24845553 11238408 20798317 25315136
24508170 30249741 21719571 17994087 21347285 29654886 28648823 15705502 15632085 2109311 6084717 11156992 10731132 12537572 10731137 2503416 11294610 17550304 15972463 29187976 8601488 8889526 25830018 24495485 25348373 8533095 9359576 18362917 19820115 24845553 11238408 20798317 25315136
EMBL
AF229609
AAF61569.1
BABH01019098
BABH01019099
BABH01019100
BABH01019101
+ More
BABH01019102 BABH01019103 BABH01019104 BABH01019105 AB445460 BAH03523.1 EU722903 ACD99701.1 AGBW02011105 OWR47210.1 KM667971 AJQ21779.1 KQ459472 KPJ00092.1 KQ461175 KPJ07919.1 MH172526 QAA95933.1 CAQQ02175586 CAQQ02175587 KK107293 EZA53360.1 QOIP01000005 RLU22270.1 GL888393 EGI61708.1 KQ976870 KYN07740.1 KQ982194 KYQ59053.1 CH479197 EDW27841.1 KQ981208 KYN44888.1 ADTU01015824 KQ976737 KYM75916.1 LC368084 BBC77943.1 KQ979763 KYN19244.1 KQ414727 KOC62606.1 GBYB01000323 JAG70090.1 AJWK01004725 AJWK01004726 AJWK01004727 AJWK01004728 GBYB01000322 JAG70089.1 CH940672 KRF77745.1 EDW57384.1 CP012528 ALC48197.1 MH105806 AWC08636.1 NNAY01000361 OXU28894.1 CM000366 EDX16959.1 AB191788 BAD54748.1 KF828804 AIN44121.1 KU376474 ANO53447.1 CH933814 EDW05654.1 OUUW01000015 SPP88490.1 SPP88489.1 KQ435713 KOX79246.1 CH379064 EDY72595.1 KRT06475.1 X51749 X02974 AB028139 AE014298 AL133506 X76202 CH954180 EDV45682.1 CH902625 EDV34941.1 CM000162 KRK06073.1 AM887687 CAP09074.1 EDX01323.1 DQ335250 ABC69732.1 MF908522 AXK92644.1 U64875 AAB06578.1 AH012033 GBXI01016111 AAN38825.1 JAC98180.1 CH916370 EDV99695.1 GAMC01015788 JAB90767.1 GAKP01017893 GAKP01017891 JAC41061.1 X89933 GDHF01008584 JAI43730.1 U97104 AAC61893.1 GDHF01033402 GDHF01005555 JAI18912.1 JAI46759.1 CH964239 EDW81927.1 AF422804 AAL56571.1 AH011253 AF442746 KQ971372 AAL40947.1 EFA09849.1 KK852575 KDR20987.1 AF318275 AF318276 AAK21872.1 GL452135 EFN77859.1 KA645686 AFP60315.1 CCAG010008024 JXJN01008099 LBMM01003652 KMQ93279.1
BABH01019102 BABH01019103 BABH01019104 BABH01019105 AB445460 BAH03523.1 EU722903 ACD99701.1 AGBW02011105 OWR47210.1 KM667971 AJQ21779.1 KQ459472 KPJ00092.1 KQ461175 KPJ07919.1 MH172526 QAA95933.1 CAQQ02175586 CAQQ02175587 KK107293 EZA53360.1 QOIP01000005 RLU22270.1 GL888393 EGI61708.1 KQ976870 KYN07740.1 KQ982194 KYQ59053.1 CH479197 EDW27841.1 KQ981208 KYN44888.1 ADTU01015824 KQ976737 KYM75916.1 LC368084 BBC77943.1 KQ979763 KYN19244.1 KQ414727 KOC62606.1 GBYB01000323 JAG70090.1 AJWK01004725 AJWK01004726 AJWK01004727 AJWK01004728 GBYB01000322 JAG70089.1 CH940672 KRF77745.1 EDW57384.1 CP012528 ALC48197.1 MH105806 AWC08636.1 NNAY01000361 OXU28894.1 CM000366 EDX16959.1 AB191788 BAD54748.1 KF828804 AIN44121.1 KU376474 ANO53447.1 CH933814 EDW05654.1 OUUW01000015 SPP88490.1 SPP88489.1 KQ435713 KOX79246.1 CH379064 EDY72595.1 KRT06475.1 X51749 X02974 AB028139 AE014298 AL133506 X76202 CH954180 EDV45682.1 CH902625 EDV34941.1 CM000162 KRK06073.1 AM887687 CAP09074.1 EDX01323.1 DQ335250 ABC69732.1 MF908522 AXK92644.1 U64875 AAB06578.1 AH012033 GBXI01016111 AAN38825.1 JAC98180.1 CH916370 EDV99695.1 GAMC01015788 JAB90767.1 GAKP01017893 GAKP01017891 JAC41061.1 X89933 GDHF01008584 JAI43730.1 U97104 AAC61893.1 GDHF01033402 GDHF01005555 JAI18912.1 JAI46759.1 CH964239 EDW81927.1 AF422804 AAL56571.1 AH011253 AF442746 KQ971372 AAL40947.1 EFA09849.1 KK852575 KDR20987.1 AF318275 AF318276 AAK21872.1 GL452135 EFN77859.1 KA645686 AFP60315.1 CCAG010008024 JXJN01008099 LBMM01003652 KMQ93279.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000015102
UP000053097
+ More
UP000279307 UP000007755 UP000078542 UP000075809 UP000008744 UP000078541 UP000005205 UP000078540 UP000078492 UP000053825 UP000092461 UP000008792 UP000092553 UP000215335 UP000000304 UP000192221 UP000009192 UP000268350 UP000053105 UP000001819 UP000000803 UP000008711 UP000007801 UP000002282 UP000001070 UP000007798 UP000095300 UP000007266 UP000027135 UP000008237 UP000095301 UP000005203 UP000091820 UP000092444 UP000092443 UP000092460 UP000078200 UP000092445 UP000036403
UP000279307 UP000007755 UP000078542 UP000075809 UP000008744 UP000078541 UP000005205 UP000078540 UP000078492 UP000053825 UP000092461 UP000008792 UP000092553 UP000215335 UP000000304 UP000192221 UP000009192 UP000268350 UP000053105 UP000001819 UP000000803 UP000008711 UP000007801 UP000002282 UP000001070 UP000007798 UP000095300 UP000007266 UP000027135 UP000008237 UP000095301 UP000005203 UP000091820 UP000092444 UP000092443 UP000092460 UP000078200 UP000092445 UP000036403
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR013525 ABC_2_trans
IPR005284 Pigment_permease/Abcg
IPR003593 AAA+_ATPase
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR003113 PI3K_adapt-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR025110 AMP-bd_C
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR013525 ABC_2_trans
IPR005284 Pigment_permease/Abcg
IPR003593 AAA+_ATPase
IPR000341 PI3K_Ras-bd_dom
IPR042236 PI3K_accessory_sf
IPR002420 PI3K_C2_dom
IPR011009 Kinase-like_dom_sf
IPR000403 PI3/4_kinase_cat_dom
IPR018936 PI3/4_kinase_CS
IPR003113 PI3K_adapt-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR036940 PI3/4_kinase_cat_sf
IPR015433 PI_Kinase
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR001263 PI3K_accessory_dom
IPR025110 AMP-bd_C
Gene 3D
ProteinModelPortal
Q9NH94
H9J087
B7XCV5
B3GQX5
A0A212F0E3
A0A0C5PTD7
+ More
A0A194Q3I4 A0A194QRF1 A0A3R5WTC8 T1GGV8 A0A026WBC3 A0A3L8DP96 F4WVN8 A0A195D4B1 A0A151XF99 B4GYI5 A0A195FWS3 A0A158NHH2 A0A195AUR0 A0A2Z5ZBX4 A0A195E2Y4 A0A0L7QVP7 A0A0C9PHK2 A0A1B0CB12 A0A0C9QIB5 A0A0Q9W770 B4MG61 A0A0M4ETI1 A0A2S0X9Z2 A0A232FDM6 B4R3U7 Q60FC5 A0A158V6Z1 A0A1W4VF76 A0A1B0YWI7 B4L879 A0A3B0KIY5 A0A3B0KPD8 A0A0M9A9I2 B5DLT3 A0A0R3NZL1 P10090 B3NTX4 B3MWF6 A0A0R1EA29 A8E1U1 B4Q0V6 Q2LD54 A0A346A6M2 Q94960 Q8IS30 B4JJE3 W8AX98 A0A034VCM1 Q17320 A0A0K8VYU6 O77423 A0A0K8W6U9 B4NDV8 A0A1I8P5B8 Q8WRR1 Q8WRF2 A0A067RAU0 Q9BH97 E2C2K3 A0A1I8MED8 T1PA82 A0A087ZW42 A0A1A9W968 A0A1B0G2T2 A0A1A9XVI0 A0A1B0B3W9 A0A1A9V0K8 A0A1B0A8G3 A0A0J7KSS0
A0A194Q3I4 A0A194QRF1 A0A3R5WTC8 T1GGV8 A0A026WBC3 A0A3L8DP96 F4WVN8 A0A195D4B1 A0A151XF99 B4GYI5 A0A195FWS3 A0A158NHH2 A0A195AUR0 A0A2Z5ZBX4 A0A195E2Y4 A0A0L7QVP7 A0A0C9PHK2 A0A1B0CB12 A0A0C9QIB5 A0A0Q9W770 B4MG61 A0A0M4ETI1 A0A2S0X9Z2 A0A232FDM6 B4R3U7 Q60FC5 A0A158V6Z1 A0A1W4VF76 A0A1B0YWI7 B4L879 A0A3B0KIY5 A0A3B0KPD8 A0A0M9A9I2 B5DLT3 A0A0R3NZL1 P10090 B3NTX4 B3MWF6 A0A0R1EA29 A8E1U1 B4Q0V6 Q2LD54 A0A346A6M2 Q94960 Q8IS30 B4JJE3 W8AX98 A0A034VCM1 Q17320 A0A0K8VYU6 O77423 A0A0K8W6U9 B4NDV8 A0A1I8P5B8 Q8WRR1 Q8WRF2 A0A067RAU0 Q9BH97 E2C2K3 A0A1I8MED8 T1PA82 A0A087ZW42 A0A1A9W968 A0A1B0G2T2 A0A1A9XVI0 A0A1B0B3W9 A0A1A9V0K8 A0A1B0A8G3 A0A0J7KSS0
Ontologies
GO
GO:0005524
GO:0055085
GO:0016021
GO:0016887
GO:0046854
GO:0016301
GO:0048015
GO:0008558
GO:1905948
GO:0007613
GO:0015842
GO:0042401
GO:0051615
GO:0031410
GO:0005275
GO:0008049
GO:0048072
GO:0042441
GO:0005395
GO:0070731
GO:0042332
GO:0098793
GO:0031409
GO:0006727
GO:0006856
GO:0090740
GO:0048770
GO:0042626
GO:0005886
PANTHER
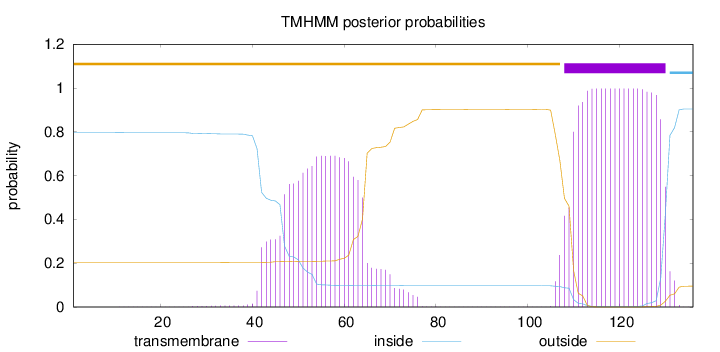
Topology
Subcellular location
Membrane
Cytoplasmic granule membrane
Cytoplasmic granule membrane
Length:
136
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.88819
Exp number, first 60 AAs:
10.5983
Total prob of N-in:
0.79650
POSSIBLE N-term signal
sequence
outside
1 - 107
TMhelix
108 - 130
inside
131 - 136
Population Genetic Test Statistics
Pi
239.316235
Theta
178.562763
Tajima's D
0.911509
CLR
0
CSRT
0.633168341582921
Interpretation
Uncertain
