Gene
KWMTBOMO05895
Pre Gene Modal
BGIBMGA002929
Annotation
PREDICTED:_protoheme_IX_farnesyltransferase?_mitochondrial_[Bombyx_mori]
Full name
Protoheme IX farnesyltransferase, mitochondrial
+ More
Adenosylhomocysteinase
Structural maintenance of chromosomes protein
Adenosylhomocysteinase
Structural maintenance of chromosomes protein
Alternative Name
Heme O synthase
Location in the cell
PlasmaMembrane Reliability : 4.373
Sequence
CDS
ATGTCATATTTAAAGGTGATACACTGTACCTTGTGCCTGAAACATGGATACGTGGGAGTTTACTCGAAACCAGTTTTATTTAAAATTACTAATAACACACTCTTCCAGCATCTTGCCAGAATATCAACAGCACAAAACTGGAGAAGTAAAGTTCCACTTAAAACTCAGACAACTGCAACGGTAAAGAATAAAGTCACCCAAGACACCAGAGTATGGAAGGAAACACCATCATATGACAGGAAGAGCAATACGGGACAGTATTGTTTGATGCTATCTAAATCACGGTTAACATCATTAGTTGTGTTGACGTCTATGGCAGGATACGCACTAGCACCTGCCCCGTTTCAACTTACTACCTTTGCATTCTGCGCTGTGGGTACTGGTCTAGTATCAGCTGCAGCAAACTCTATAAATCAGTATCATGAAGTACCATTTGATGCCCAAATGTCGAGGACAAAGAACAGGGTGCTTGTCAAAGGTTTATTAGAGCCCGTCCATGCAATAGGTTTTGCAGCATTGACAAGCAGCGTAGGCCTTGGCGTTTTGTATTTTGGGGTCAACCCCTTGACAGCTGCACTAGGAGCAGGAAACCTCATCCTCTACACCTCAGTGTACACTCCAATGAAAAGAATGTCCATACTTAACACCTGGCTTGGCTCTGTAGTTGGAGCCATACCCCCCTTGATGGGATGGGCGGGCTGTACCGGGGGACTGGACGCTGGCGCGCTGGTGCTGGGCGCCTTGCTCTACTCGTGGCAGTTTCCACACTTCAACGCGCTCTCTTGGAATTTGCGACCAGACTACTCGCGGGCCGGGTATCGAATGATGGCCGTCACTAATCCCGCACTATGCCGAAGGGTTGCGCTTAGACACACAGGAATAATCACAGCAACATGCCTGGTATCACCTTATATAGGCGTTACAAACACGTGGTTCGCACTTGAATCATTACCATTGAATATTTATTTTATGTATTTAGCATGGCAATTCTACCAAAAATCAGACAGCGCAAGTTCGCGGAAGCTATTCAGATTTTCTCTACTTCACCTACCAGCATTAATGTTGCTAATGCTAGTAAATAAGAAGTACTGGAGTTCGAGTGACACTCAAGAGCAGACCGACACCAAAATCCAGGACGTAGGCATATTAGACAAGAGAATTACAGTACTACCTCGAGGACCGGTCGTTGTGGCCCAAGGAAACGACAAACAGTGCTAA
Protein
MSYLKVIHCTLCLKHGYVGVYSKPVLFKITNNTLFQHLARISTAQNWRSKVPLKTQTTATVKNKVTQDTRVWKETPSYDRKSNTGQYCLMLSKSRLTSLVVLTSMAGYALAPAPFQLTTFAFCAVGTGLVSAAANSINQYHEVPFDAQMSRTKNRVLVKGLLEPVHAIGFAALTSSVGLGVLYFGVNPLTAALGAGNLILYTSVYTPMKRMSILNTWLGSVVGAIPPLMGWAGCTGGLDAGALVLGALLYSWQFPHFNALSWNLRPDYSRAGYRMMAVTNPALCRRVALRHTGIITATCLVSPYIGVTNTWFALESLPLNIYFMYLAWQFYQKSDSASSRKLFRFSLLHLPALMLLMLVNKKYWSSSDTQEQTDTKIQDVGILDKRITVLPRGPVVVAQGNDKQC
Summary
Description
Converts protoheme IX and farnesyl diphosphate to heme O.
Catalytic Activity
H2O + S-adenosyl-L-homocysteine = adenosine + L-homocysteine
Cofactor
NAD(+)
Similarity
Belongs to the ubiA prenyltransferase family.
Belongs to the adenosylhomocysteinase family.
Belongs to the SMC family.
Belongs to the adenosylhomocysteinase family.
Belongs to the SMC family.
Uniprot
A0A194Q844
A0A194QWG6
A0A212EKQ8
A0A2H1WZH3
A0A2W1BK11
A0A1B6DZP8
+ More
A0A336M9Y1 A0A1B6MME4 B4LRA0 A0A2S2QA70 A0A1B6JVW7 B4JPM9 B4Q8U2 B4NZA1 B3N976 A0A2S2NAR1 Q9VKZ1 B3MPR6 C4WVL8 X1WJJ8 A0A0K8TKQ5 A0A0P4VL84 A0A1W4UG35 A0A1B6GJ74 B4KJK0 A0A1J1HTJ9 A0A0Q9XGJ0 A0A0A1WJ35 A0A3Q0ITB6 A0A3Q0IMS8 A0A1I8MZW5 A0A139WBV5 B0WQ76 W8BZA2 A0A1I8PHB2 A0A1I8PH37 E2BI52 A0A1Y1MTJ7 Q17L26 A0A182H5U9 A0A1B0CSN5 A0A0A9Y1Q1 A0A0K8T0H1 A0A023ESP8 A0A0L7QSL8 A0A1L8DZE2 A0A0K8V5P4 U4UQS3 A0A3B0K7S5 A0A1Q3FGH6 B4NQQ9 N6TCT9 K7IRR7 A0A195CBD9 A0A1A9Y6Y1 A0A1A9UT82 A0A026WWR5 A0A1B0BD28 A0A1B0DH26 E9JB06 A0A1A9ZVN7 D3TLZ7 A0A088A3H8 Q29CJ3 A0A2J7Q212 A0A2A3E3B8 V9IMH2 A0A151WRV5 A0A182M2F1 F4W874 B4G792 A0A182P674 A0A182RUP9 A0A182LMY2 A0A158NQG6 A0A182IVC0 A0A182Q0L9 A0A154NYN4 A0A182I777 W5JCA3 A0A2M4ABX0 A0A182JSP7 A0A182XDC4 A0A084VYN8 A0A182F5S5 A0A1S4H085 A0A195DUL9 A0A2M4BK45 Q7PWQ5 A0A2M4BK01 A0A2M4BK20 A0A182URC7 T1DQE7 A0A2M4ACG3 A0A2M3Z0W1 A0A2M4AEV6
A0A336M9Y1 A0A1B6MME4 B4LRA0 A0A2S2QA70 A0A1B6JVW7 B4JPM9 B4Q8U2 B4NZA1 B3N976 A0A2S2NAR1 Q9VKZ1 B3MPR6 C4WVL8 X1WJJ8 A0A0K8TKQ5 A0A0P4VL84 A0A1W4UG35 A0A1B6GJ74 B4KJK0 A0A1J1HTJ9 A0A0Q9XGJ0 A0A0A1WJ35 A0A3Q0ITB6 A0A3Q0IMS8 A0A1I8MZW5 A0A139WBV5 B0WQ76 W8BZA2 A0A1I8PHB2 A0A1I8PH37 E2BI52 A0A1Y1MTJ7 Q17L26 A0A182H5U9 A0A1B0CSN5 A0A0A9Y1Q1 A0A0K8T0H1 A0A023ESP8 A0A0L7QSL8 A0A1L8DZE2 A0A0K8V5P4 U4UQS3 A0A3B0K7S5 A0A1Q3FGH6 B4NQQ9 N6TCT9 K7IRR7 A0A195CBD9 A0A1A9Y6Y1 A0A1A9UT82 A0A026WWR5 A0A1B0BD28 A0A1B0DH26 E9JB06 A0A1A9ZVN7 D3TLZ7 A0A088A3H8 Q29CJ3 A0A2J7Q212 A0A2A3E3B8 V9IMH2 A0A151WRV5 A0A182M2F1 F4W874 B4G792 A0A182P674 A0A182RUP9 A0A182LMY2 A0A158NQG6 A0A182IVC0 A0A182Q0L9 A0A154NYN4 A0A182I777 W5JCA3 A0A2M4ABX0 A0A182JSP7 A0A182XDC4 A0A084VYN8 A0A182F5S5 A0A1S4H085 A0A195DUL9 A0A2M4BK45 Q7PWQ5 A0A2M4BK01 A0A2M4BK20 A0A182URC7 T1DQE7 A0A2M4ACG3 A0A2M3Z0W1 A0A2M4AEV6
EC Number
2.5.1.-
3.3.1.1
3.3.1.1
Pubmed
26354079
22118469
28756777
17994087
22936249
17550304
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 27129103 25830018 25315136 18362917 19820115 24495485 20798317 28004739 17510324 26483478 25401762 26823975 24945155 23537049 20075255 24508170 21282665 20353571 15632085 21719571 20966253 21347285 20920257 23761445 24438588 12364791
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 27129103 25830018 25315136 18362917 19820115 24495485 20798317 28004739 17510324 26483478 25401762 26823975 24945155 23537049 20075255 24508170 21282665 20353571 15632085 21719571 20966253 21347285 20920257 23761445 24438588 12364791
EMBL
KQ459302
KPJ01698.1
KQ461175
KPJ07911.1
AGBW02014216
OWR42079.1
+ More
ODYU01012212 SOQ58402.1 KZ149998 PZC75379.1 GEDC01006167 JAS31131.1 UFQT01000735 SSX26830.1 GEBQ01019669 GEBQ01002867 JAT20308.1 JAT37110.1 CH940649 EDW64570.2 GGMS01005318 MBY74521.1 GECU01004387 JAT03320.1 CH916372 EDV98859.1 CM000361 CM002910 EDX04507.1 KMY89492.1 CM000157 EDW88796.1 CH954177 EDV58511.1 GGMR01001606 MBY14225.1 AE014134 AY058639 AAF52915.1 AAL13868.1 CH902620 EDV32314.1 AK341579 BAH71938.1 ABLF02017103 GDAI01002651 JAI14952.1 GDKW01003457 JAI53138.1 GECZ01026209 GECZ01015673 GECZ01007298 JAS43560.1 JAS54096.1 JAS62471.1 CH933807 EDW13580.1 CVRI01000020 CRK90708.1 KRG03886.1 GBXI01015611 JAC98680.1 KQ971372 KYB25345.1 DS232036 EDS32736.1 GAMC01001913 JAC04643.1 GL448450 EFN84623.1 GEZM01024410 JAV87795.1 CH477219 EAT47401.1 JXUM01026687 KQ560764 KXJ80973.1 AJWK01026312 GBHO01018043 JAG25561.1 GBRD01006915 GDHC01013747 JAG58906.1 JAQ04882.1 GAPW01001295 JAC12303.1 KQ414758 KOC61461.1 GFDF01002280 JAV11804.1 GDHF01018087 GDHF01008834 JAI34227.1 JAI43480.1 KB632326 ERL92400.1 OUUW01000006 SPP81686.1 GFDL01008387 JAV26658.1 CH964295 EDW86698.2 APGK01034968 KB740923 ENN78114.1 KQ978023 KYM98015.1 KK107078 EZA60502.1 JXJN01012270 AJVK01015316 GL770707 EFZ09997.1 EZ422449 ADD18725.1 CH475462 EAL29304.3 NEVH01019374 PNF22622.1 KZ288403 PBC26205.1 JR052111 AEY61624.1 KQ982803 KYQ50548.1 AXCM01000336 GL887898 EGI69569.1 CH479180 EDW28346.1 ADTU01023315 ADTU01023316 AXCN02002086 KQ434783 KZC04733.1 APCN01003368 ADMH02001840 ETN60953.1 GGFK01004938 MBW38259.1 ATLV01018382 KE525231 KFB43082.1 AAAB01008984 KQ980322 KYN16573.1 GGFJ01004212 MBW53353.1 EAA14890.4 GGFJ01004211 MBW53352.1 GGFJ01004213 MBW53354.1 GAMD01001200 JAB00391.1 GGFK01005150 MBW38471.1 GGFM01001327 MBW22078.1 GGFK01005993 MBW39314.1
ODYU01012212 SOQ58402.1 KZ149998 PZC75379.1 GEDC01006167 JAS31131.1 UFQT01000735 SSX26830.1 GEBQ01019669 GEBQ01002867 JAT20308.1 JAT37110.1 CH940649 EDW64570.2 GGMS01005318 MBY74521.1 GECU01004387 JAT03320.1 CH916372 EDV98859.1 CM000361 CM002910 EDX04507.1 KMY89492.1 CM000157 EDW88796.1 CH954177 EDV58511.1 GGMR01001606 MBY14225.1 AE014134 AY058639 AAF52915.1 AAL13868.1 CH902620 EDV32314.1 AK341579 BAH71938.1 ABLF02017103 GDAI01002651 JAI14952.1 GDKW01003457 JAI53138.1 GECZ01026209 GECZ01015673 GECZ01007298 JAS43560.1 JAS54096.1 JAS62471.1 CH933807 EDW13580.1 CVRI01000020 CRK90708.1 KRG03886.1 GBXI01015611 JAC98680.1 KQ971372 KYB25345.1 DS232036 EDS32736.1 GAMC01001913 JAC04643.1 GL448450 EFN84623.1 GEZM01024410 JAV87795.1 CH477219 EAT47401.1 JXUM01026687 KQ560764 KXJ80973.1 AJWK01026312 GBHO01018043 JAG25561.1 GBRD01006915 GDHC01013747 JAG58906.1 JAQ04882.1 GAPW01001295 JAC12303.1 KQ414758 KOC61461.1 GFDF01002280 JAV11804.1 GDHF01018087 GDHF01008834 JAI34227.1 JAI43480.1 KB632326 ERL92400.1 OUUW01000006 SPP81686.1 GFDL01008387 JAV26658.1 CH964295 EDW86698.2 APGK01034968 KB740923 ENN78114.1 KQ978023 KYM98015.1 KK107078 EZA60502.1 JXJN01012270 AJVK01015316 GL770707 EFZ09997.1 EZ422449 ADD18725.1 CH475462 EAL29304.3 NEVH01019374 PNF22622.1 KZ288403 PBC26205.1 JR052111 AEY61624.1 KQ982803 KYQ50548.1 AXCM01000336 GL887898 EGI69569.1 CH479180 EDW28346.1 ADTU01023315 ADTU01023316 AXCN02002086 KQ434783 KZC04733.1 APCN01003368 ADMH02001840 ETN60953.1 GGFK01004938 MBW38259.1 ATLV01018382 KE525231 KFB43082.1 AAAB01008984 KQ980322 KYN16573.1 GGFJ01004212 MBW53353.1 EAA14890.4 GGFJ01004211 MBW53352.1 GGFJ01004213 MBW53354.1 GAMD01001200 JAB00391.1 GGFK01005150 MBW38471.1 GGFM01001327 MBW22078.1 GGFK01005993 MBW39314.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000008792
UP000001070
UP000000304
+ More
UP000002282 UP000008711 UP000000803 UP000007801 UP000007819 UP000192221 UP000009192 UP000183832 UP000079169 UP000095301 UP000007266 UP000002320 UP000095300 UP000008237 UP000008820 UP000069940 UP000249989 UP000092461 UP000053825 UP000030742 UP000268350 UP000007798 UP000019118 UP000002358 UP000078542 UP000092443 UP000078200 UP000053097 UP000092460 UP000092462 UP000092445 UP000005203 UP000001819 UP000235965 UP000242457 UP000075809 UP000075883 UP000007755 UP000008744 UP000075885 UP000075900 UP000075882 UP000005205 UP000075880 UP000075886 UP000076502 UP000075840 UP000000673 UP000075881 UP000076407 UP000030765 UP000069272 UP000078492 UP000007062 UP000075903
UP000002282 UP000008711 UP000000803 UP000007801 UP000007819 UP000192221 UP000009192 UP000183832 UP000079169 UP000095301 UP000007266 UP000002320 UP000095300 UP000008237 UP000008820 UP000069940 UP000249989 UP000092461 UP000053825 UP000030742 UP000268350 UP000007798 UP000019118 UP000002358 UP000078542 UP000092443 UP000078200 UP000053097 UP000092460 UP000092462 UP000092445 UP000005203 UP000001819 UP000235965 UP000242457 UP000075809 UP000075883 UP000007755 UP000008744 UP000075885 UP000075900 UP000075882 UP000005205 UP000075880 UP000075886 UP000076502 UP000075840 UP000000673 UP000075881 UP000076407 UP000030765 UP000069272 UP000078492 UP000007062 UP000075903
PRIDE
Pfam
Interpro
IPR006369
Protohaem_IX_farnesylTrfase
+ More
IPR000537 UbiA_prenyltransferase
IPR016315 Protohaem_IX_farnesylTrfase_mt
IPR030470 UbiA_prenylTrfase_CS
IPR000043 Adenosylhomocysteinase-like
IPR015878 Ado_hCys_hydrolase_NAD-bd
IPR042172 Adenosylhomocyst_ase-like_sf
IPR034373 Adenosylhomocysteinase
IPR020082 S-Ado-L-homoCys_hydrolase_CS
IPR036291 NAD(P)-bd_dom_sf
IPR017970 Homeobox_CS
IPR001356 Homeobox_dom
IPR009057 Homeobox-like_sf
IPR036277 SMC_hinge_sf
IPR003395 RecF/RecN/SMC_N
IPR010935 SMC_hinge
IPR024704 SMC
IPR027120 Smc2_ABC
IPR027417 P-loop_NTPase
IPR000537 UbiA_prenyltransferase
IPR016315 Protohaem_IX_farnesylTrfase_mt
IPR030470 UbiA_prenylTrfase_CS
IPR000043 Adenosylhomocysteinase-like
IPR015878 Ado_hCys_hydrolase_NAD-bd
IPR042172 Adenosylhomocyst_ase-like_sf
IPR034373 Adenosylhomocysteinase
IPR020082 S-Ado-L-homoCys_hydrolase_CS
IPR036291 NAD(P)-bd_dom_sf
IPR017970 Homeobox_CS
IPR001356 Homeobox_dom
IPR009057 Homeobox-like_sf
IPR036277 SMC_hinge_sf
IPR003395 RecF/RecN/SMC_N
IPR010935 SMC_hinge
IPR024704 SMC
IPR027120 Smc2_ABC
IPR027417 P-loop_NTPase
Gene 3D
ProteinModelPortal
A0A194Q844
A0A194QWG6
A0A212EKQ8
A0A2H1WZH3
A0A2W1BK11
A0A1B6DZP8
+ More
A0A336M9Y1 A0A1B6MME4 B4LRA0 A0A2S2QA70 A0A1B6JVW7 B4JPM9 B4Q8U2 B4NZA1 B3N976 A0A2S2NAR1 Q9VKZ1 B3MPR6 C4WVL8 X1WJJ8 A0A0K8TKQ5 A0A0P4VL84 A0A1W4UG35 A0A1B6GJ74 B4KJK0 A0A1J1HTJ9 A0A0Q9XGJ0 A0A0A1WJ35 A0A3Q0ITB6 A0A3Q0IMS8 A0A1I8MZW5 A0A139WBV5 B0WQ76 W8BZA2 A0A1I8PHB2 A0A1I8PH37 E2BI52 A0A1Y1MTJ7 Q17L26 A0A182H5U9 A0A1B0CSN5 A0A0A9Y1Q1 A0A0K8T0H1 A0A023ESP8 A0A0L7QSL8 A0A1L8DZE2 A0A0K8V5P4 U4UQS3 A0A3B0K7S5 A0A1Q3FGH6 B4NQQ9 N6TCT9 K7IRR7 A0A195CBD9 A0A1A9Y6Y1 A0A1A9UT82 A0A026WWR5 A0A1B0BD28 A0A1B0DH26 E9JB06 A0A1A9ZVN7 D3TLZ7 A0A088A3H8 Q29CJ3 A0A2J7Q212 A0A2A3E3B8 V9IMH2 A0A151WRV5 A0A182M2F1 F4W874 B4G792 A0A182P674 A0A182RUP9 A0A182LMY2 A0A158NQG6 A0A182IVC0 A0A182Q0L9 A0A154NYN4 A0A182I777 W5JCA3 A0A2M4ABX0 A0A182JSP7 A0A182XDC4 A0A084VYN8 A0A182F5S5 A0A1S4H085 A0A195DUL9 A0A2M4BK45 Q7PWQ5 A0A2M4BK01 A0A2M4BK20 A0A182URC7 T1DQE7 A0A2M4ACG3 A0A2M3Z0W1 A0A2M4AEV6
A0A336M9Y1 A0A1B6MME4 B4LRA0 A0A2S2QA70 A0A1B6JVW7 B4JPM9 B4Q8U2 B4NZA1 B3N976 A0A2S2NAR1 Q9VKZ1 B3MPR6 C4WVL8 X1WJJ8 A0A0K8TKQ5 A0A0P4VL84 A0A1W4UG35 A0A1B6GJ74 B4KJK0 A0A1J1HTJ9 A0A0Q9XGJ0 A0A0A1WJ35 A0A3Q0ITB6 A0A3Q0IMS8 A0A1I8MZW5 A0A139WBV5 B0WQ76 W8BZA2 A0A1I8PHB2 A0A1I8PH37 E2BI52 A0A1Y1MTJ7 Q17L26 A0A182H5U9 A0A1B0CSN5 A0A0A9Y1Q1 A0A0K8T0H1 A0A023ESP8 A0A0L7QSL8 A0A1L8DZE2 A0A0K8V5P4 U4UQS3 A0A3B0K7S5 A0A1Q3FGH6 B4NQQ9 N6TCT9 K7IRR7 A0A195CBD9 A0A1A9Y6Y1 A0A1A9UT82 A0A026WWR5 A0A1B0BD28 A0A1B0DH26 E9JB06 A0A1A9ZVN7 D3TLZ7 A0A088A3H8 Q29CJ3 A0A2J7Q212 A0A2A3E3B8 V9IMH2 A0A151WRV5 A0A182M2F1 F4W874 B4G792 A0A182P674 A0A182RUP9 A0A182LMY2 A0A158NQG6 A0A182IVC0 A0A182Q0L9 A0A154NYN4 A0A182I777 W5JCA3 A0A2M4ABX0 A0A182JSP7 A0A182XDC4 A0A084VYN8 A0A182F5S5 A0A1S4H085 A0A195DUL9 A0A2M4BK45 Q7PWQ5 A0A2M4BK01 A0A2M4BK20 A0A182URC7 T1DQE7 A0A2M4ACG3 A0A2M3Z0W1 A0A2M4AEV6
Ontologies
PATHWAY
GO
GO:0016021
GO:0008495
GO:0048034
GO:0031966
GO:0070069
GO:1902600
GO:0022900
GO:0006783
GO:0008535
GO:0016491
GO:0019510
GO:0004013
GO:0006730
GO:0051287
GO:0016740
GO:0005739
GO:0005634
GO:0045333
GO:0043565
GO:0006355
GO:0005694
GO:0051276
GO:0005524
GO:0016765
GO:0016818
GO:0003676
GO:0008026
GO:0016597
Topology
Subcellular location
Mitochondrion membrane
Nucleus
Nucleus
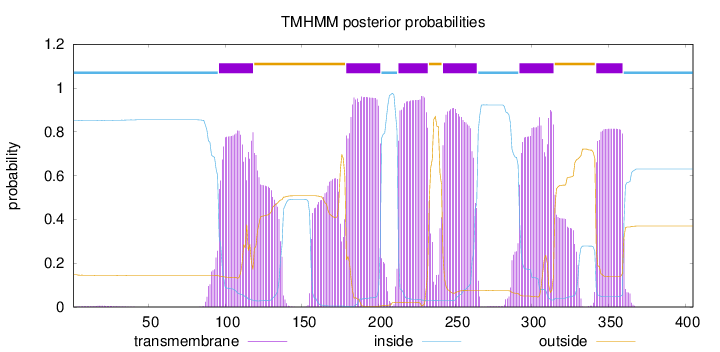
Length:
405
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
141.81474
Exp number, first 60 AAs:
0.10948
Total prob of N-in:
0.85282
inside
1 - 95
TMhelix
96 - 118
outside
119 - 178
TMhelix
179 - 201
inside
202 - 212
TMhelix
213 - 232
outside
233 - 241
TMhelix
242 - 264
inside
265 - 291
TMhelix
292 - 314
outside
315 - 341
TMhelix
342 - 359
inside
360 - 405
Population Genetic Test Statistics
Pi
212.230269
Theta
168.80409
Tajima's D
0.374697
CLR
0.045442
CSRT
0.476026198690066
Interpretation
Uncertain
