Pre Gene Modal
BGIBMGA002779
Annotation
PREDICTED:_FACT_complex_subunit_spt16_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.932
Sequence
CDS
ATGTCTAATATATCATTGGACAAAGAGACGTTTTATCGTCGCATGAAAAAGCTGTATGCGACCTGGAAGGCTGTAGCTTCAGATCCTAAGAGTGATGATGCCCTGTCAAAGGTGGATTGCTTAGTGTCGTGTGTAGGAGTAGATGAAGAGACACTCTACAGCAAATCAACATCACTTCAGACATGGCTGTTTGGATATGAGTTACCTGACACAATCACTGTCTTAACAGAGCATAGCATGTGTTTTTTAGCCAGTAAGAAGAAGATTGAATTCCTGAGACAAATAGAAAATGGTAAAGATGAAACTGAATTGCCCCCAGCAAAACTTCTTATTAGAGACAGAAATGACAAAGACAAGGAAAACTTCAACAAACTTCTCCAAGAAATAAAGAAATCAAAATCAGGCAAGACACTGGGTATTTTCGTGAAAGACAATTATCCAGGTGAATTTTGTGAAAGTTGGAAAGCAGTTCTGAAGGGAGAGAAGTTTGAAAATGTCGATGTCAGTTCAGCGATTGCATTATTAATGGCGCCAAAAGAAGACTCTGAAATTATTACAATAAAAAAAGCATGTCTGGTCACAGTGGATGTCTTCACAAAATACTTAAAAGATCAGATAATGGAAATTATTGATTCAGATAAGAAAGTCAAACATTCAAAACTTGCTGAAGGTGTGGAAACAGCAGTTTCAGATAAGAAATATGTAACAGGAGTCGATACAAGTCAAGTTGACATGTGTTATCCTCCTATTATACAGTCAGGCGGACACTATAGTTTGAAATTCAGTGCAGTATCTGATAAAAACCACTTACACTTTGGCGCCATAGTATGTTCACTAGGTGCTAGATACAAATCATATTGTTCGAACATAGTCCGTACATTACTGGTCAACCCAACTGATGAAGTACAAAGTAATTACAATTTCTTGTTAAATATTGAAGAAGAGGTGATGAAGAGTCTTGTAGCTGGAGCGAAATTATCTACAGTTTATGAAGCTGGTCTAGCGTTAGCCAAGAAAGAGAAACCTAACTTAGTTGAAAATCTCACTAAGAGTTTTGGATTCGCTATGGGTATAGAATTTCGTGAAAGTTCCATTATTATTGGACCTAAAACAAATGTTACCGCGAAAAAAGGAATGGTCTTCAACATAAACATTGGCTTGGCGAACTTAACAAACAGTAATGCCTCAGATAAGGAGGGGAAGACATACGCGCTCTTCATTGGTGACACAGTATTAGTTAACGAAGAGCAGCCAGCGTCTCTACTAACACAGTCTAAGAAAAAAGTAAAAAATATAGGAATATTCTTGAAAGATGACGATGAAGAAGAAGAAGAGGAGAAAGAAAACAAGACTGAAATACTCGAAAATGGAGGTAGAGGCAAAAGGACTGCAGTGATTGAATCGAAATTGAGAACAGAGCATTCATCTGAGGAAAAACGTAAGGAGCATCAGCGTGAACTTGCTATATCTTTGAATGAAAAGGCCAAAGAAAGATTAGCCAAACAGTCGACCGGTAAAGATACAGAAAAACTAAGGAAAAGCACAGTTTCATACAAGAGTATTAGTCAGATGCCACGTGAGAATGAAGTTAAAGAATTAAAATTATATGTCGATCGTAAATACGAAACAGTAATCCTGCCAATATTCGGTGTCCCAGTACCATTCCATATATCTACAATTAAGAACATCTCGCAGTCAGTGGAAGGCGATTACACGTACCTTAGAATTAACTTTTTCCATCCCGGCGCCACTATGGGCAGGAATGAAGGCGGGAATTACTCTCAACCGGACGCCACCTTCGTTAAGGAAGTTACATACCGAAGTACAAACACAAAAGAACCGGGTGAGATATCGCCGCCTTCGTCAAACTTGAACACTGGATTTAGACTTATTAAAGAAGTACAGAAAAAATTCAAAACCAGAGAAGCAGAAGAACGTGAAAAGGAAGATTTGGTCAAACAGGACACGTTAATATTATCACAGAACAAAGGAAATCCTAAGTTAAAAGATTTGTACATACGTCCCAATATTGTTACGAAGAGAATGAGTGGTTCATTAGAAGCTCACACAAACGGCTTTAGATTCACATCGGTACGAGGCGATAAAGTGGACATTTTGTACAATAATATAAAGAATGCATTCTTTCAACCTTGTGATGGAGAAATGATTATACTTTTACATTTCCATTTGAAACATGCCATTATGTTTGGTAAAAAAAAGCATGTGGACGTGCAATTTTACACCGAAGTCGGTGAGATAACCACAGACTTAGGAAAGCATCAACACATGCACGACCGCGACGACCTCGCCGCCGAGCAGAGCGAACGGGAGCTGAGACACAAACTGAAGGTGGCCTTCAAGAGCTTCTGTGAACGAGTCGAGAACATGACCAAGCAAGAAGTCGAGTTCGATACACCTTTCAGAGAGCTTGGTTTCCCTGGAGCTCCATATCGGAGTACAGTTTTACTGCAGCCTACCTCCGGAGCCCTTGTCAATCTTACTGAATGGCCACCATTCGTGATTGCATTAGAAGATGTAGAGCTCGTCCACTTTGAGAGAGTACAATTTCACCTCAAAAATTTCGATATGGTCTTCGTATTTAAGGACTACGCCAAGAAAGTGGCCATGGTTAATGCCGTGCCTATGGACATGCTGGATCATGTCAAAGAATGGTTGAATTCTTGTGATATCCGGTACTCGGAGGGTATACAATCGTTGAACTGGACCAAAGTTATGAAAACTATAACAGATGACATTGAGGGCTTCTTCGAAAATGGTGGCTGGTCATTTTTGGATCCAGAATCTGATGGTGAAAATGAACAAGAGGAGGAAGATAGTGAAGATGAAGATGAAGCGTACGAGCCTACGGACGCTGAGTCTGATGAATCTGAAGGGTCTGACTATGATTCTGAGGCGTCTGATGAATCTGACGCTTCGGGTGATAGTGAAGAGGAAGACGAAGAATCCGGTAAAGACTGGTCAGATTTAGAGCGGGAAGCTGCCGAAGAGGATAAAAAAGACCGCAACTTCGATCGAGATGACTTCGACCGGAAACGTAAAGGACGAAACCAACAACGCTCGTACGACTCTGATGAGGGAGCCGCCAAGAAAAGCAAACATGACAAGTCTAATCATAAAAGTTCTAGTTCAAACCACAAAAGCTCTAGTTCCAATCATAAAAGTTCGAGCTCAAACCATAAGAGCTCCAGTCATAAAAGCCCTTCAAAGAACAGTAGCAGCCCTTCCAAGAACCGGGATAAGCACCACAAATCATCCCACCATGATCGACACCACAAGAGTGACCATAAGTCCTCTAGTAAAGACAAAGACAAGAAACATTCTAGTGACCATAAATCACACAAACGGTCACGCGATGACAGCAGAGAACACGGCAACAGCGCAAAGAAAGCGAAAAAATAA
Protein
MSNISLDKETFYRRMKKLYATWKAVASDPKSDDALSKVDCLVSCVGVDEETLYSKSTSLQTWLFGYELPDTITVLTEHSMCFLASKKKIEFLRQIENGKDETELPPAKLLIRDRNDKDKENFNKLLQEIKKSKSGKTLGIFVKDNYPGEFCESWKAVLKGEKFENVDVSSAIALLMAPKEDSEIITIKKACLVTVDVFTKYLKDQIMEIIDSDKKVKHSKLAEGVETAVSDKKYVTGVDTSQVDMCYPPIIQSGGHYSLKFSAVSDKNHLHFGAIVCSLGARYKSYCSNIVRTLLVNPTDEVQSNYNFLLNIEEEVMKSLVAGAKLSTVYEAGLALAKKEKPNLVENLTKSFGFAMGIEFRESSIIIGPKTNVTAKKGMVFNINIGLANLTNSNASDKEGKTYALFIGDTVLVNEEQPASLLTQSKKKVKNIGIFLKDDDEEEEEEKENKTEILENGGRGKRTAVIESKLRTEHSSEEKRKEHQRELAISLNEKAKERLAKQSTGKDTEKLRKSTVSYKSISQMPRENEVKELKLYVDRKYETVILPIFGVPVPFHISTIKNISQSVEGDYTYLRINFFHPGATMGRNEGGNYSQPDATFVKEVTYRSTNTKEPGEISPPSSNLNTGFRLIKEVQKKFKTREAEEREKEDLVKQDTLILSQNKGNPKLKDLYIRPNIVTKRMSGSLEAHTNGFRFTSVRGDKVDILYNNIKNAFFQPCDGEMIILLHFHLKHAIMFGKKKHVDVQFYTEVGEITTDLGKHQHMHDRDDLAAEQSERELRHKLKVAFKSFCERVENMTKQEVEFDTPFRELGFPGAPYRSTVLLQPTSGALVNLTEWPPFVIALEDVELVHFERVQFHLKNFDMVFVFKDYAKKVAMVNAVPMDMLDHVKEWLNSCDIRYSEGIQSLNWTKVMKTITDDIEGFFENGGWSFLDPESDGENEQEEEDSEDEDEAYEPTDAESDESEGSDYDSEASDESDASGDSEEEDEESGKDWSDLEREAAEEDKKDRNFDRDDFDRKRKGRNQQRSYDSDEGAAKKSKHDKSNHKSSSSNHKSSSSNHKSSSSNHKSSSHKSPSKNSSSPSKNRDKHHKSSHHDRHHKSDHKSSSKDKDKKHSSDHKSHKRSRDDSREHGNSAKKAKK
Summary
Uniprot
H9IZU4
A0A2W1BKG3
A0A2H1WZC8
S4PD03
A0A212EKS6
A0A194Q8H6
+ More
A0A195DVD0 A0A2A3EQP3 A0A088A0L4 A0A151XE56 A0A158NJY0 A0A026X2Y7 F4WIL1 A0A1Y1MRU8 A0A154NYX3 A0A0T6AVK8 A0A195BIX5 A0A1L8E2L1 E9IMH2 U5EVZ2 A0A1J1IVB2 A0A0C9QZW5 A0A0C9RGX3 A0A194QSF1 D6WL41 A0A1J1IVI5 A0A195F4I6 A0A1W4WHF4 A0A067RH63 K7IQB7 A0A232EZS7 A0A2J7QBI7 A0A2J7QBH9 W5J3B2 A0A084WMU2 A0A1S4FUK7 A0A182FFL0 A0A3F2Z0E0 A0A336MRE7 Q16NS9 A0A2M4BBH1 A0A182N7E1 A0A3F2Z0F0 A0A182HBI3 A0A1Q3G0N4 A0A2M4ABW8 A0A182L3Y1 Q7PTA1 A0A182QXD3 A0A182PIU2 A0A1Q3EXI0 A0A182Y868 A0A182K770 A0A336KYU7 A0A182M4N1 A0A182J7G2 A0A182R6R6 A0A1B6DZ32 A0A1B6E447 A0A2D1QUC8 A0A182WI16 A0A0L7QX46 E2BWZ8 T1PB37 A0A0A1XQE4 A0A0A1WXK8 T1PKD5 A0A1I8Q6A9 A0A0L0BV73 A0A1I8Q6E2 A0A1A9Z2U3 A0A1A9YN61 A0A0K8WGK9 A0A1A9UJ99 A0A0K8US82 A0A034VLW7 A0A0K8U443 A0A0K8VDU8 A0A1B0FNC0 A0A182X293 A0A034VMM6 A0A1B6H1F3 A0A1B0AY34 A0A1A9WSA1 A0A182I6F4 B4LDE4 A0A0Q9WGW6 N6U723 B4L9P2 A0A0J7L219 B4IZY3 B4GS33 A0A0Q9XV52 B5DPP2 W8C5I4 A0A3L8DLE4 A0A0R3P3V8 A0A0M4ECL5 B3M787 T1I5C1
A0A195DVD0 A0A2A3EQP3 A0A088A0L4 A0A151XE56 A0A158NJY0 A0A026X2Y7 F4WIL1 A0A1Y1MRU8 A0A154NYX3 A0A0T6AVK8 A0A195BIX5 A0A1L8E2L1 E9IMH2 U5EVZ2 A0A1J1IVB2 A0A0C9QZW5 A0A0C9RGX3 A0A194QSF1 D6WL41 A0A1J1IVI5 A0A195F4I6 A0A1W4WHF4 A0A067RH63 K7IQB7 A0A232EZS7 A0A2J7QBI7 A0A2J7QBH9 W5J3B2 A0A084WMU2 A0A1S4FUK7 A0A182FFL0 A0A3F2Z0E0 A0A336MRE7 Q16NS9 A0A2M4BBH1 A0A182N7E1 A0A3F2Z0F0 A0A182HBI3 A0A1Q3G0N4 A0A2M4ABW8 A0A182L3Y1 Q7PTA1 A0A182QXD3 A0A182PIU2 A0A1Q3EXI0 A0A182Y868 A0A182K770 A0A336KYU7 A0A182M4N1 A0A182J7G2 A0A182R6R6 A0A1B6DZ32 A0A1B6E447 A0A2D1QUC8 A0A182WI16 A0A0L7QX46 E2BWZ8 T1PB37 A0A0A1XQE4 A0A0A1WXK8 T1PKD5 A0A1I8Q6A9 A0A0L0BV73 A0A1I8Q6E2 A0A1A9Z2U3 A0A1A9YN61 A0A0K8WGK9 A0A1A9UJ99 A0A0K8US82 A0A034VLW7 A0A0K8U443 A0A0K8VDU8 A0A1B0FNC0 A0A182X293 A0A034VMM6 A0A1B6H1F3 A0A1B0AY34 A0A1A9WSA1 A0A182I6F4 B4LDE4 A0A0Q9WGW6 N6U723 B4L9P2 A0A0J7L219 B4IZY3 B4GS33 A0A0Q9XV52 B5DPP2 W8C5I4 A0A3L8DLE4 A0A0R3P3V8 A0A0M4ECL5 B3M787 T1I5C1
Pubmed
19121390
28756777
23622113
22118469
26354079
21347285
+ More
24508170 21719571 28004739 21282665 18362917 19820115 24845553 20075255 28648823 20920257 23761445 24438588 17510324 26483478 20966253 12364791 14747013 17210077 25244985 20798317 25315136 25830018 26108605 25348373 17994087 23537049 15632085 24495485 30249741
24508170 21719571 28004739 21282665 18362917 19820115 24845553 20075255 28648823 20920257 23761445 24438588 17510324 26483478 20966253 12364791 14747013 17210077 25244985 20798317 25315136 25830018 26108605 25348373 17994087 23537049 15632085 24495485 30249741
EMBL
BABH01019158
BABH01019159
BABH01019160
KZ149998
PZC75378.1
ODYU01012212
+ More
SOQ58401.1 GAIX01005197 JAA87363.1 AGBW02014216 OWR42080.1 KQ459302 KPJ01699.1 KQ980304 KYN16801.1 KZ288201 PBC33546.1 KQ982254 KYQ58639.1 ADTU01018467 ADTU01018468 KK107020 EZA62602.1 GL888176 EGI65947.1 GEZM01023411 GEZM01023410 JAV88403.1 KQ434783 KZC04875.1 LJIG01022714 KRT79094.1 KQ976465 KYM84289.1 GFDF01001126 JAV12958.1 GL764146 EFZ18248.1 GANO01000785 JAB59086.1 CVRI01000061 CRL04111.1 GBYB01006232 JAG75999.1 GBYB01006231 JAG75998.1 KQ461175 KPJ07910.1 KQ971343 EFA04061.1 CRL04112.1 KQ981820 KYN35296.1 KK852473 KDR23156.1 NNAY01001504 OXU23767.1 NEVH01016298 PNF25942.1 PNF25941.1 ADMH02002207 ETN57813.1 ATLV01024512 KE525352 KFB51536.1 UFQT01002094 SSX32650.1 CH477810 EAT36003.1 GGFJ01001190 MBW50331.1 JXUM01124927 KQ566886 KXJ69752.1 GFDL01001719 JAV33326.1 GGFK01004963 MBW38284.1 AAAB01008807 EAA04225.5 AXCN02000568 GFDL01015032 JAV20013.1 UFQS01001174 UFQT01001174 SSX09388.1 SSX29290.1 AXCM01007401 GEDC01006355 JAS30943.1 GEDC01004596 JAS32702.1 KY285063 ATP16166.1 KQ414705 KOC63193.1 GL451188 EFN79862.1 KA645178 AFP59807.1 GBXI01001115 JAD13177.1 GBXI01010936 JAD03356.1 KA649221 AFP63850.1 JRES01001294 KNC23873.1 GDHF01002243 GDHF01002043 JAI50071.1 JAI50271.1 GDHF01023114 GDHF01009431 JAI29200.1 JAI42883.1 GAKP01016173 GAKP01016172 JAC42780.1 GDHF01030991 GDHF01024209 JAI21323.1 JAI28105.1 GDHF01015273 GDHF01002719 JAI37041.1 JAI49595.1 CCAG010000676 GAKP01016170 GAKP01016169 JAC42783.1 GECZ01001255 JAS68514.1 JXJN01005529 APCN01003635 CH940647 EDW68882.1 KRF84060.1 APGK01037730 APGK01037731 KB740948 KB632364 ENN77450.1 ERL93546.1 CH933816 EDW17090.1 LBMM01001250 KMQ96643.1 CH916366 EDV96755.1 CH479188 EDW40568.1 KRG07603.1 CH379069 EDY73564.1 GAMC01009021 JAB97534.1 QOIP01000006 RLU21260.1 KRT07854.1 CP012525 ALC43184.1 CH902618 EDV38748.1 ACPB03022602 ACPB03022603
SOQ58401.1 GAIX01005197 JAA87363.1 AGBW02014216 OWR42080.1 KQ459302 KPJ01699.1 KQ980304 KYN16801.1 KZ288201 PBC33546.1 KQ982254 KYQ58639.1 ADTU01018467 ADTU01018468 KK107020 EZA62602.1 GL888176 EGI65947.1 GEZM01023411 GEZM01023410 JAV88403.1 KQ434783 KZC04875.1 LJIG01022714 KRT79094.1 KQ976465 KYM84289.1 GFDF01001126 JAV12958.1 GL764146 EFZ18248.1 GANO01000785 JAB59086.1 CVRI01000061 CRL04111.1 GBYB01006232 JAG75999.1 GBYB01006231 JAG75998.1 KQ461175 KPJ07910.1 KQ971343 EFA04061.1 CRL04112.1 KQ981820 KYN35296.1 KK852473 KDR23156.1 NNAY01001504 OXU23767.1 NEVH01016298 PNF25942.1 PNF25941.1 ADMH02002207 ETN57813.1 ATLV01024512 KE525352 KFB51536.1 UFQT01002094 SSX32650.1 CH477810 EAT36003.1 GGFJ01001190 MBW50331.1 JXUM01124927 KQ566886 KXJ69752.1 GFDL01001719 JAV33326.1 GGFK01004963 MBW38284.1 AAAB01008807 EAA04225.5 AXCN02000568 GFDL01015032 JAV20013.1 UFQS01001174 UFQT01001174 SSX09388.1 SSX29290.1 AXCM01007401 GEDC01006355 JAS30943.1 GEDC01004596 JAS32702.1 KY285063 ATP16166.1 KQ414705 KOC63193.1 GL451188 EFN79862.1 KA645178 AFP59807.1 GBXI01001115 JAD13177.1 GBXI01010936 JAD03356.1 KA649221 AFP63850.1 JRES01001294 KNC23873.1 GDHF01002243 GDHF01002043 JAI50071.1 JAI50271.1 GDHF01023114 GDHF01009431 JAI29200.1 JAI42883.1 GAKP01016173 GAKP01016172 JAC42780.1 GDHF01030991 GDHF01024209 JAI21323.1 JAI28105.1 GDHF01015273 GDHF01002719 JAI37041.1 JAI49595.1 CCAG010000676 GAKP01016170 GAKP01016169 JAC42783.1 GECZ01001255 JAS68514.1 JXJN01005529 APCN01003635 CH940647 EDW68882.1 KRF84060.1 APGK01037730 APGK01037731 KB740948 KB632364 ENN77450.1 ERL93546.1 CH933816 EDW17090.1 LBMM01001250 KMQ96643.1 CH916366 EDV96755.1 CH479188 EDW40568.1 KRG07603.1 CH379069 EDY73564.1 GAMC01009021 JAB97534.1 QOIP01000006 RLU21260.1 KRT07854.1 CP012525 ALC43184.1 CH902618 EDV38748.1 ACPB03022602 ACPB03022603
Proteomes
UP000005204
UP000007151
UP000053268
UP000078492
UP000242457
UP000005203
+ More
UP000075809 UP000005205 UP000053097 UP000007755 UP000076502 UP000078540 UP000183832 UP000053240 UP000007266 UP000078541 UP000192223 UP000027135 UP000002358 UP000215335 UP000235965 UP000000673 UP000030765 UP000069272 UP000075903 UP000008820 UP000075884 UP000069940 UP000249989 UP000075882 UP000007062 UP000075886 UP000075885 UP000076408 UP000075881 UP000075883 UP000075880 UP000075900 UP000075920 UP000053825 UP000008237 UP000095301 UP000095300 UP000037069 UP000092445 UP000092443 UP000078200 UP000092444 UP000076407 UP000092460 UP000091820 UP000075840 UP000008792 UP000019118 UP000030742 UP000009192 UP000036403 UP000001070 UP000008744 UP000001819 UP000279307 UP000092553 UP000007801 UP000015103
UP000075809 UP000005205 UP000053097 UP000007755 UP000076502 UP000078540 UP000183832 UP000053240 UP000007266 UP000078541 UP000192223 UP000027135 UP000002358 UP000215335 UP000235965 UP000000673 UP000030765 UP000069272 UP000075903 UP000008820 UP000075884 UP000069940 UP000249989 UP000075882 UP000007062 UP000075886 UP000075885 UP000076408 UP000075881 UP000075883 UP000075880 UP000075900 UP000075920 UP000053825 UP000008237 UP000095301 UP000095300 UP000037069 UP000092445 UP000092443 UP000078200 UP000092444 UP000076407 UP000092460 UP000091820 UP000075840 UP000008792 UP000019118 UP000030742 UP000009192 UP000036403 UP000001070 UP000008744 UP000001819 UP000279307 UP000092553 UP000007801 UP000015103
PRIDE
Pfam
Interpro
IPR033825
Spt16_M24
+ More
IPR029148 FACT-Spt16_Nlobe
IPR011993 PH-like_dom_sf
IPR013953 FACT_Spt16
IPR029149 Creatin/AminoP/Spt16_NTD
IPR036005 Creatinase/aminopeptidase-like
IPR040258 Spt16
IPR013719 DUF1747
IPR000994 Pept_M24
IPR027417 P-loop_NTPase
IPR020588 RecA_ATP-bd
IPR013632 DNA_recomb/repair_Rad51_C
IPR027806 HARBI1_dom
IPR029148 FACT-Spt16_Nlobe
IPR011993 PH-like_dom_sf
IPR013953 FACT_Spt16
IPR029149 Creatin/AminoP/Spt16_NTD
IPR036005 Creatinase/aminopeptidase-like
IPR040258 Spt16
IPR013719 DUF1747
IPR000994 Pept_M24
IPR027417 P-loop_NTPase
IPR020588 RecA_ATP-bd
IPR013632 DNA_recomb/repair_Rad51_C
IPR027806 HARBI1_dom
Gene 3D
ProteinModelPortal
H9IZU4
A0A2W1BKG3
A0A2H1WZC8
S4PD03
A0A212EKS6
A0A194Q8H6
+ More
A0A195DVD0 A0A2A3EQP3 A0A088A0L4 A0A151XE56 A0A158NJY0 A0A026X2Y7 F4WIL1 A0A1Y1MRU8 A0A154NYX3 A0A0T6AVK8 A0A195BIX5 A0A1L8E2L1 E9IMH2 U5EVZ2 A0A1J1IVB2 A0A0C9QZW5 A0A0C9RGX3 A0A194QSF1 D6WL41 A0A1J1IVI5 A0A195F4I6 A0A1W4WHF4 A0A067RH63 K7IQB7 A0A232EZS7 A0A2J7QBI7 A0A2J7QBH9 W5J3B2 A0A084WMU2 A0A1S4FUK7 A0A182FFL0 A0A3F2Z0E0 A0A336MRE7 Q16NS9 A0A2M4BBH1 A0A182N7E1 A0A3F2Z0F0 A0A182HBI3 A0A1Q3G0N4 A0A2M4ABW8 A0A182L3Y1 Q7PTA1 A0A182QXD3 A0A182PIU2 A0A1Q3EXI0 A0A182Y868 A0A182K770 A0A336KYU7 A0A182M4N1 A0A182J7G2 A0A182R6R6 A0A1B6DZ32 A0A1B6E447 A0A2D1QUC8 A0A182WI16 A0A0L7QX46 E2BWZ8 T1PB37 A0A0A1XQE4 A0A0A1WXK8 T1PKD5 A0A1I8Q6A9 A0A0L0BV73 A0A1I8Q6E2 A0A1A9Z2U3 A0A1A9YN61 A0A0K8WGK9 A0A1A9UJ99 A0A0K8US82 A0A034VLW7 A0A0K8U443 A0A0K8VDU8 A0A1B0FNC0 A0A182X293 A0A034VMM6 A0A1B6H1F3 A0A1B0AY34 A0A1A9WSA1 A0A182I6F4 B4LDE4 A0A0Q9WGW6 N6U723 B4L9P2 A0A0J7L219 B4IZY3 B4GS33 A0A0Q9XV52 B5DPP2 W8C5I4 A0A3L8DLE4 A0A0R3P3V8 A0A0M4ECL5 B3M787 T1I5C1
A0A195DVD0 A0A2A3EQP3 A0A088A0L4 A0A151XE56 A0A158NJY0 A0A026X2Y7 F4WIL1 A0A1Y1MRU8 A0A154NYX3 A0A0T6AVK8 A0A195BIX5 A0A1L8E2L1 E9IMH2 U5EVZ2 A0A1J1IVB2 A0A0C9QZW5 A0A0C9RGX3 A0A194QSF1 D6WL41 A0A1J1IVI5 A0A195F4I6 A0A1W4WHF4 A0A067RH63 K7IQB7 A0A232EZS7 A0A2J7QBI7 A0A2J7QBH9 W5J3B2 A0A084WMU2 A0A1S4FUK7 A0A182FFL0 A0A3F2Z0E0 A0A336MRE7 Q16NS9 A0A2M4BBH1 A0A182N7E1 A0A3F2Z0F0 A0A182HBI3 A0A1Q3G0N4 A0A2M4ABW8 A0A182L3Y1 Q7PTA1 A0A182QXD3 A0A182PIU2 A0A1Q3EXI0 A0A182Y868 A0A182K770 A0A336KYU7 A0A182M4N1 A0A182J7G2 A0A182R6R6 A0A1B6DZ32 A0A1B6E447 A0A2D1QUC8 A0A182WI16 A0A0L7QX46 E2BWZ8 T1PB37 A0A0A1XQE4 A0A0A1WXK8 T1PKD5 A0A1I8Q6A9 A0A0L0BV73 A0A1I8Q6E2 A0A1A9Z2U3 A0A1A9YN61 A0A0K8WGK9 A0A1A9UJ99 A0A0K8US82 A0A034VLW7 A0A0K8U443 A0A0K8VDU8 A0A1B0FNC0 A0A182X293 A0A034VMM6 A0A1B6H1F3 A0A1B0AY34 A0A1A9WSA1 A0A182I6F4 B4LDE4 A0A0Q9WGW6 N6U723 B4L9P2 A0A0J7L219 B4IZY3 B4GS33 A0A0Q9XV52 B5DPP2 W8C5I4 A0A3L8DLE4 A0A0R3P3V8 A0A0M4ECL5 B3M787 T1I5C1
PDB
4Z2N
E-value=3.68686e-137,
Score=1255
Ontologies
GO
PANTHER
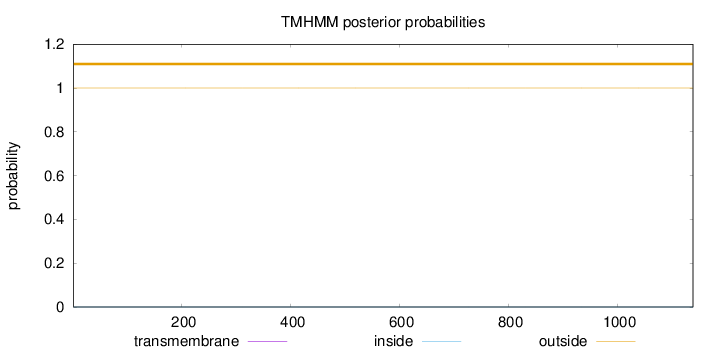
Topology
Length:
1139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00319
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00015
outside
1 - 1139
Population Genetic Test Statistics
Pi
212.906001
Theta
181.07814
Tajima's D
0.631728
CLR
0.371382
CSRT
0.549672516374181
Interpretation
Uncertain
