Gene
KWMTBOMO05892
Pre Gene Modal
BGIBMGA002930
Annotation
PREDICTED:_uncharacterized_protein_LOC106718540_[Papilio_machaon]
Full name
TRPL translocation defect protein 14
Location in the cell
Nuclear Reliability : 1.959
Sequence
CDS
ATGGCCAATCAATCTCAGAAAATCGTTTACAAGCTGGTTCTTACTGGAGGACCCTGCGGCGGCAAAACAACGGGACAGTCACGTTTAAGTACCTTCTTCGAGAACCTAGGATGGAAGGTGTTCCGGGTGCCGGAAACGGCTACCGTCCTTCTCAGCGGAGGCATAAAGTTTGCAGACCTCAGTCCTGAAGAAGCCTCGAAATTCCAAGAGAACCTATTGAAAACGATGATACAGATTGAGAACACGTTCTTCGAACTCGGCAGAACTAGTCAGAGGAATTGTCTGATAATCTGCGATCGAGGAGCCATGGACGCTAGTGCATTTATATCGAAGGAGAAATGGGAGGCGATACTCGCATCTAACAACTGGAATAATGTCGAACTACGCGACAACAGATACAACCACATCATGCATATGGTATCCGCTGCTAACGGAGCTGAAGACTTCTATTCCACGGAGGATCATGCTTGCCGATCTGAAGGAGTGGAACTCGCCAGAGAGCTGGACTACAAGGCGGCCGCCGCTTGGATCGGTCACCCGTACTTCGACGTGATCGACAATTCGACAGATTTCGATAAAAAGATGAATCGTTTGATCGCGTGCGTCTGCCAGAAGATCGGCATCGATACCGGCGACCGGCTCAACGTCAACTCGAAGAAGCGCAAGTTCCTCATCAAGGCGCCTCTCGCTCCCGACTCGGAGTTCCCGCCCTTCCAAGACTTCGACGTCGTTCACAACTACCTGCAGAGCGATTTGCGGAAAGCACAAGTCCGGCTACGTAAACGCGGACAGAAGGGACACTGGTCGTACATACACACGTTAAGGAAGTTCCACCCCACCAACGGACAGTCCGTCGAGGTCCGCACGCAACTGACGCACCGAGATTATTTGAATCTGTTGCCGCAGCGCGATGACGCTCATCTGACCATCTTCAAGAAGCGTCGTTGTTTCATCTACAACAACCAGTACTATCAGTTGGACATTTACAGGCAACCGACGCATCCCAGATGTCGCGGTTTGGTTCTCCTAGAAACCTACAGCGCTGCTTACGATCAGACCATTTTGCTGGCTTCATTACCCAAGTTTCTGACCATTGAGAAAGAAGTCACCGGAGATCCAGCATATTCAATGTACAATCTGTCATTGAAAGAAGATTGGAAGACCTCGAAAAGGTACTACGCTGGCACTGAGTGCAATGGTCACACTAAGTTGACCAACGGTCATGGCAACGTCAAAATTTGCCATTCAAAGATGAACGGTCATAGTGAGGAGAAAATCGGGCTCGCCTGTGACAAAATCAACGGTTGCGAGAAATCTGAAGTCAGGAAAACCAACGGTCACCATCACGTCAACGGTCACGGCGTGAAAACCGATCGAAACGGTATCACGAGAAACGGTAACGCCCACACCAATGAACTGCAAGACATCAAACAAAGAATTGATCTCGGGTCGCCAAAGATTCAAAAGGTCGCCGTAAAGATTTAA
Protein
MANQSQKIVYKLVLTGGPCGGKTTGQSRLSTFFENLGWKVFRVPETATVLLSGGIKFADLSPEEASKFQENLLKTMIQIENTFFELGRTSQRNCLIICDRGAMDASAFISKEKWEAILASNNWNNVELRDNRYNHIMHMVSAANGAEDFYSTEDHACRSEGVELARELDYKAAAAWIGHPYFDVIDNSTDFDKKMNRLIACVCQKIGIDTGDRLNVNSKKRKFLIKAPLAPDSEFPPFQDFDVVHNYLQSDLRKAQVRLRKRGQKGHWSYIHTLRKFHPTNGQSVEVRTQLTHRDYLNLLPQRDDAHLTIFKKRRCFIYNNQYYQLDIYRQPTHPRCRGLVLLETYSAAYDQTILLASLPKFLTIEKEVTGDPAYSMYNLSLKEDWKTSKRYYAGTECNGHTKLTNGHGNVKICHSKMNGHSEEKIGLACDKINGCEKSEVRKTNGHHHVNGHGVKTDRNGITRNGNAHTNELQDIKQRIDLGSPKIQKVAVKI
Summary
Description
GTP-binding protein which is required for the light-dependent internalization of the TRPL ion channel from the rhabdomere on the apical surface of photoreceptor cells to the cell body and for the recycling of TRPL back to the rhabdomere in the dark. Binds to 3-phosphoinositide (PtdIns(3)P) and phosphatic acid and so may interact with membranes, particularly the early endosome membrane where PtdIns(3)P is predominantly localized. Plays a role in preventing photoreceptor degeneration.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
GTP-binding
Lipid-binding
Nucleotide-binding
Protein transport
Reference proteome
Transport
Feature
chain TRPL translocation defect protein 14
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J095
A0A3S2PLE6
A0A2W1BRT0
A0A194Q9L1
A0A194QQX5
A0A2H1WZ88
+ More
A0A1E1WKB5 A0A0L7LGI3 A0A212EKQ7 A0A2A4J7V0 K7IVI6 V9IKM3 A0A0C9REA7 A0A158NF37 W8BQC4 F4WEK1 E2B3I4 A0A3L8E577 A0A0A1WZ18 A0A069DZ00 A0A224XP27 A0A0V0G872 A0A023F8F4 W8C8C8 A0A0P4VUQ1 A0A0K8VM66 A0A1B6MCV5 A0A034VQH4 A0A146KNE8 A0A0A9XJH2 U5ESV8 A0A1I8NZV2 A0A1B6CCI7 A0A1B6EQH2 A0A2J7R7P3 A0A088AVP1 A0A3B0JR84 Q7K556-2 N6VF79 A0A0K8WLJ6 A0A3B0JXP7 Q7K556 A0A1B6MK29 A0A0Q9WVZ4 A0A0J9RFM9 A0A1I8NHI9 B4NMY7 T1P9X6 A0A0N8P025 A0A0J9RFV7 A0A1B6F8A9 A0A1I8NZY2 A0A0P8Y2A9 A0A0R1DXL7 A0A336LYL4 A0A0Q5VKX2 A0A336LUP4 T1E1Z0 W8BCP0 A0A0R1DXK9 A0A0Q5VZN6 A0A0K8TNM6 Q7PPH7 A0A023ESP0 A0A1B6KRJ5 A0A1Q3FY97 A0A1Q3FY70 D6X271 E2AE06 A0A1B6F1F5 A0A0A1X3A5 A0A0R3NV51 A0A151WPS7 A0A195FMB3 A0A146LYV1 A0A2M4A876 A0A1B0FKW4 A0A0Q9WIN4 A0A3F2YQ92 A0A1B6LAB5 E0VTI1 A0A2M4A8D5 A0A1A9UXB5 A0A2M3Z122 A0A2M4A8T5 A0A0Q9WGD4 T1DP48 A0A195C438 A0A2M4CJV5 A0A1B6ELE5 A0A0K8VN37 A0A3F2YQA0 A0A2M4BML9 A0A2M4BNH6 A0A1A9WBI6 A0A034VTY1 A0A232F5X0 A0A0K8UUD6 A0A2M4CVE4
A0A1E1WKB5 A0A0L7LGI3 A0A212EKQ7 A0A2A4J7V0 K7IVI6 V9IKM3 A0A0C9REA7 A0A158NF37 W8BQC4 F4WEK1 E2B3I4 A0A3L8E577 A0A0A1WZ18 A0A069DZ00 A0A224XP27 A0A0V0G872 A0A023F8F4 W8C8C8 A0A0P4VUQ1 A0A0K8VM66 A0A1B6MCV5 A0A034VQH4 A0A146KNE8 A0A0A9XJH2 U5ESV8 A0A1I8NZV2 A0A1B6CCI7 A0A1B6EQH2 A0A2J7R7P3 A0A088AVP1 A0A3B0JR84 Q7K556-2 N6VF79 A0A0K8WLJ6 A0A3B0JXP7 Q7K556 A0A1B6MK29 A0A0Q9WVZ4 A0A0J9RFM9 A0A1I8NHI9 B4NMY7 T1P9X6 A0A0N8P025 A0A0J9RFV7 A0A1B6F8A9 A0A1I8NZY2 A0A0P8Y2A9 A0A0R1DXL7 A0A336LYL4 A0A0Q5VKX2 A0A336LUP4 T1E1Z0 W8BCP0 A0A0R1DXK9 A0A0Q5VZN6 A0A0K8TNM6 Q7PPH7 A0A023ESP0 A0A1B6KRJ5 A0A1Q3FY97 A0A1Q3FY70 D6X271 E2AE06 A0A1B6F1F5 A0A0A1X3A5 A0A0R3NV51 A0A151WPS7 A0A195FMB3 A0A146LYV1 A0A2M4A876 A0A1B0FKW4 A0A0Q9WIN4 A0A3F2YQ92 A0A1B6LAB5 E0VTI1 A0A2M4A8D5 A0A1A9UXB5 A0A2M3Z122 A0A2M4A8T5 A0A0Q9WGD4 T1DP48 A0A195C438 A0A2M4CJV5 A0A1B6ELE5 A0A0K8VN37 A0A3F2YQA0 A0A2M4BML9 A0A2M4BNH6 A0A1A9WBI6 A0A034VTY1 A0A232F5X0 A0A0K8UUD6 A0A2M4CVE4
Pubmed
19121390
28756777
26354079
26227816
22118469
20075255
+ More
21347285 24495485 21719571 20798317 30249741 25830018 26334808 25474469 27129103 25348373 26823975 25401762 10731132 12537572 12537569 26509977 15632085 17994087 22936249 25315136 17550304 24330624 26369729 12364791 24945155 18362917 19820115 20566863 28648823
21347285 24495485 21719571 20798317 30249741 25830018 26334808 25474469 27129103 25348373 26823975 25401762 10731132 12537572 12537569 26509977 15632085 17994087 22936249 25315136 17550304 24330624 26369729 12364791 24945155 18362917 19820115 20566863 28648823
EMBL
BABH01019163
RSAL01000003
RVE54786.1
KZ149998
PZC75376.1
KQ459302
+ More
KPJ01700.1 KQ461175 KPJ07908.1 ODYU01012212 SOQ58399.1 GDQN01007297 GDQN01005833 GDQN01003743 GDQN01003535 JAT83757.1 JAT85221.1 JAT87311.1 JAT87519.1 JTDY01001200 KOB74562.1 AGBW02014216 OWR42082.1 NWSH01002572 PCG67949.1 JR049709 AEY61046.1 GBYB01011497 JAG81264.1 ADTU01013727 ADTU01013728 GAMC01007547 JAB99008.1 GL888103 EGI67364.1 GL445346 EFN89736.1 QOIP01000001 RLU27515.1 GBXI01010396 GBXI01004307 JAD03896.1 JAD09985.1 GBGD01001505 JAC87384.1 GFTR01006236 JAW10190.1 GECL01001754 JAP04370.1 GBBI01001081 JAC17631.1 GAMC01007546 JAB99009.1 GDKW01003169 JAI53426.1 GDHF01012371 JAI39943.1 GEBQ01006220 JAT33757.1 GAKP01013391 JAC45561.1 GDHC01021807 JAP96821.1 GBHO01023510 GBHO01023505 JAG20094.1 JAG20099.1 GANO01004656 JAB55215.1 GEDC01026121 JAS11177.1 GECZ01029568 JAS40201.1 NEVH01006727 PNF36857.1 OUUW01000001 SPP75856.1 AE013599 AY051446 AAK92870.1 CM000071 ENO01720.2 GDHF01023757 GDHF01000352 JAI28557.1 JAI51962.1 SPP75858.1 GEBQ01003669 JAT36308.1 CH964282 KRG00272.1 CM002911 KMY94848.1 EDW85726.2 KA645532 AFP60161.1 CH902619 KPU75940.1 KMY94847.1 GECZ01023425 JAS46344.1 KPU75941.1 CM000158 KRK00002.1 UFQT01000211 SSX21753.1 CH954179 KQS62087.1 SSX21752.1 GALA01001655 JAA93197.1 GAMC01007545 JAB99010.1 KRK00001.1 KQS62088.1 GDAI01001634 JAI15969.1 AAAB01008948 EAA10383.4 GAPW01001311 JAC12287.1 GEBQ01025937 JAT14040.1 GFDL01002537 JAV32508.1 GFDL01002536 JAV32509.1 KQ971371 EFA09905.2 GL438827 EFN68239.1 GECZ01025792 JAS43977.1 GBXI01009062 JAD05230.1 KRT02659.1 KQ982854 KYQ49817.1 KQ981490 KYN41382.1 GDHC01005865 JAQ12764.1 GGFK01003676 MBW36997.1 CCAG010015818 CCAG010015819 CH940648 KRF80485.1 GEBQ01019416 JAT20561.1 DS235766 EEB16687.1 GGFK01003704 MBW37025.1 GGFM01001463 MBW22214.1 GGFK01003707 MBW37028.1 KRF80484.1 GAMD01002265 JAA99325.1 KQ978287 KYM95622.1 GGFL01001361 MBW65539.1 GECZ01031048 JAS38721.1 GDHF01032911 GDHF01012324 JAI19403.1 JAI39990.1 GGFJ01005188 MBW54329.1 GGFJ01005187 MBW54328.1 GAKP01013390 JAC45562.1 NNAY01000837 OXU26256.1 GDHF01022176 JAI30138.1 GGFL01005061 MBW69239.1
KPJ01700.1 KQ461175 KPJ07908.1 ODYU01012212 SOQ58399.1 GDQN01007297 GDQN01005833 GDQN01003743 GDQN01003535 JAT83757.1 JAT85221.1 JAT87311.1 JAT87519.1 JTDY01001200 KOB74562.1 AGBW02014216 OWR42082.1 NWSH01002572 PCG67949.1 JR049709 AEY61046.1 GBYB01011497 JAG81264.1 ADTU01013727 ADTU01013728 GAMC01007547 JAB99008.1 GL888103 EGI67364.1 GL445346 EFN89736.1 QOIP01000001 RLU27515.1 GBXI01010396 GBXI01004307 JAD03896.1 JAD09985.1 GBGD01001505 JAC87384.1 GFTR01006236 JAW10190.1 GECL01001754 JAP04370.1 GBBI01001081 JAC17631.1 GAMC01007546 JAB99009.1 GDKW01003169 JAI53426.1 GDHF01012371 JAI39943.1 GEBQ01006220 JAT33757.1 GAKP01013391 JAC45561.1 GDHC01021807 JAP96821.1 GBHO01023510 GBHO01023505 JAG20094.1 JAG20099.1 GANO01004656 JAB55215.1 GEDC01026121 JAS11177.1 GECZ01029568 JAS40201.1 NEVH01006727 PNF36857.1 OUUW01000001 SPP75856.1 AE013599 AY051446 AAK92870.1 CM000071 ENO01720.2 GDHF01023757 GDHF01000352 JAI28557.1 JAI51962.1 SPP75858.1 GEBQ01003669 JAT36308.1 CH964282 KRG00272.1 CM002911 KMY94848.1 EDW85726.2 KA645532 AFP60161.1 CH902619 KPU75940.1 KMY94847.1 GECZ01023425 JAS46344.1 KPU75941.1 CM000158 KRK00002.1 UFQT01000211 SSX21753.1 CH954179 KQS62087.1 SSX21752.1 GALA01001655 JAA93197.1 GAMC01007545 JAB99010.1 KRK00001.1 KQS62088.1 GDAI01001634 JAI15969.1 AAAB01008948 EAA10383.4 GAPW01001311 JAC12287.1 GEBQ01025937 JAT14040.1 GFDL01002537 JAV32508.1 GFDL01002536 JAV32509.1 KQ971371 EFA09905.2 GL438827 EFN68239.1 GECZ01025792 JAS43977.1 GBXI01009062 JAD05230.1 KRT02659.1 KQ982854 KYQ49817.1 KQ981490 KYN41382.1 GDHC01005865 JAQ12764.1 GGFK01003676 MBW36997.1 CCAG010015818 CCAG010015819 CH940648 KRF80485.1 GEBQ01019416 JAT20561.1 DS235766 EEB16687.1 GGFK01003704 MBW37025.1 GGFM01001463 MBW22214.1 GGFK01003707 MBW37028.1 KRF80484.1 GAMD01002265 JAA99325.1 KQ978287 KYM95622.1 GGFL01001361 MBW65539.1 GECZ01031048 JAS38721.1 GDHF01032911 GDHF01012324 JAI19403.1 JAI39990.1 GGFJ01005188 MBW54329.1 GGFJ01005187 MBW54328.1 GAKP01013390 JAC45562.1 NNAY01000837 OXU26256.1 GDHF01022176 JAI30138.1 GGFL01005061 MBW69239.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000218220 UP000002358 UP000005205 UP000007755 UP000008237 UP000279307 UP000095300 UP000235965 UP000005203 UP000268350 UP000000803 UP000001819 UP000007798 UP000095301 UP000007801 UP000002282 UP000008711 UP000007062 UP000007266 UP000000311 UP000075809 UP000078541 UP000092444 UP000008792 UP000069272 UP000009046 UP000078200 UP000078542 UP000091820 UP000215335
UP000218220 UP000002358 UP000005205 UP000007755 UP000008237 UP000279307 UP000095300 UP000235965 UP000005203 UP000268350 UP000000803 UP000001819 UP000007798 UP000095301 UP000007801 UP000002282 UP000008711 UP000007062 UP000007266 UP000000311 UP000075809 UP000078541 UP000092444 UP000008792 UP000069272 UP000009046 UP000078200 UP000078542 UP000091820 UP000215335
Interpro
Gene 3D
ProteinModelPortal
H9J095
A0A3S2PLE6
A0A2W1BRT0
A0A194Q9L1
A0A194QQX5
A0A2H1WZ88
+ More
A0A1E1WKB5 A0A0L7LGI3 A0A212EKQ7 A0A2A4J7V0 K7IVI6 V9IKM3 A0A0C9REA7 A0A158NF37 W8BQC4 F4WEK1 E2B3I4 A0A3L8E577 A0A0A1WZ18 A0A069DZ00 A0A224XP27 A0A0V0G872 A0A023F8F4 W8C8C8 A0A0P4VUQ1 A0A0K8VM66 A0A1B6MCV5 A0A034VQH4 A0A146KNE8 A0A0A9XJH2 U5ESV8 A0A1I8NZV2 A0A1B6CCI7 A0A1B6EQH2 A0A2J7R7P3 A0A088AVP1 A0A3B0JR84 Q7K556-2 N6VF79 A0A0K8WLJ6 A0A3B0JXP7 Q7K556 A0A1B6MK29 A0A0Q9WVZ4 A0A0J9RFM9 A0A1I8NHI9 B4NMY7 T1P9X6 A0A0N8P025 A0A0J9RFV7 A0A1B6F8A9 A0A1I8NZY2 A0A0P8Y2A9 A0A0R1DXL7 A0A336LYL4 A0A0Q5VKX2 A0A336LUP4 T1E1Z0 W8BCP0 A0A0R1DXK9 A0A0Q5VZN6 A0A0K8TNM6 Q7PPH7 A0A023ESP0 A0A1B6KRJ5 A0A1Q3FY97 A0A1Q3FY70 D6X271 E2AE06 A0A1B6F1F5 A0A0A1X3A5 A0A0R3NV51 A0A151WPS7 A0A195FMB3 A0A146LYV1 A0A2M4A876 A0A1B0FKW4 A0A0Q9WIN4 A0A3F2YQ92 A0A1B6LAB5 E0VTI1 A0A2M4A8D5 A0A1A9UXB5 A0A2M3Z122 A0A2M4A8T5 A0A0Q9WGD4 T1DP48 A0A195C438 A0A2M4CJV5 A0A1B6ELE5 A0A0K8VN37 A0A3F2YQA0 A0A2M4BML9 A0A2M4BNH6 A0A1A9WBI6 A0A034VTY1 A0A232F5X0 A0A0K8UUD6 A0A2M4CVE4
A0A1E1WKB5 A0A0L7LGI3 A0A212EKQ7 A0A2A4J7V0 K7IVI6 V9IKM3 A0A0C9REA7 A0A158NF37 W8BQC4 F4WEK1 E2B3I4 A0A3L8E577 A0A0A1WZ18 A0A069DZ00 A0A224XP27 A0A0V0G872 A0A023F8F4 W8C8C8 A0A0P4VUQ1 A0A0K8VM66 A0A1B6MCV5 A0A034VQH4 A0A146KNE8 A0A0A9XJH2 U5ESV8 A0A1I8NZV2 A0A1B6CCI7 A0A1B6EQH2 A0A2J7R7P3 A0A088AVP1 A0A3B0JR84 Q7K556-2 N6VF79 A0A0K8WLJ6 A0A3B0JXP7 Q7K556 A0A1B6MK29 A0A0Q9WVZ4 A0A0J9RFM9 A0A1I8NHI9 B4NMY7 T1P9X6 A0A0N8P025 A0A0J9RFV7 A0A1B6F8A9 A0A1I8NZY2 A0A0P8Y2A9 A0A0R1DXL7 A0A336LYL4 A0A0Q5VKX2 A0A336LUP4 T1E1Z0 W8BCP0 A0A0R1DXK9 A0A0Q5VZN6 A0A0K8TNM6 Q7PPH7 A0A023ESP0 A0A1B6KRJ5 A0A1Q3FY97 A0A1Q3FY70 D6X271 E2AE06 A0A1B6F1F5 A0A0A1X3A5 A0A0R3NV51 A0A151WPS7 A0A195FMB3 A0A146LYV1 A0A2M4A876 A0A1B0FKW4 A0A0Q9WIN4 A0A3F2YQ92 A0A1B6LAB5 E0VTI1 A0A2M4A8D5 A0A1A9UXB5 A0A2M3Z122 A0A2M4A8T5 A0A0Q9WGD4 T1DP48 A0A195C438 A0A2M4CJV5 A0A1B6ELE5 A0A0K8VN37 A0A3F2YQA0 A0A2M4BML9 A0A2M4BNH6 A0A1A9WBI6 A0A034VTY1 A0A232F5X0 A0A0K8UUD6 A0A2M4CVE4
Ontologies
GO
Topology
Subcellular location
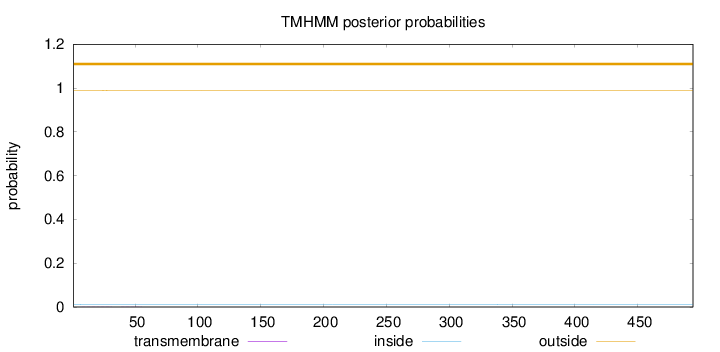
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01614
Exp number, first 60 AAs:
0.01396
Total prob of N-in:
0.01182
outside
1 - 494
Population Genetic Test Statistics
Pi
209.195739
Theta
171.074664
Tajima's D
0.688537
CLR
0.409937
CSRT
0.574671266436678
Interpretation
Uncertain
