Gene
KWMTBOMO05888
Annotation
PREDICTED:_NADPH_oxidase_4-like_[Bombyx_mori]
Full name
NADPH oxidase 4
Alternative Name
Kidney oxidase-1
Kidney superoxide-producing NADPH oxidase
Renal NAD(P)H-oxidase
Kidney superoxide-producing NADPH oxidase
Renal NAD(P)H-oxidase
Location in the cell
PlasmaMembrane Reliability : 4.959
Sequence
CDS
ATGAAAATAAAGACGTATCTTTGCGGCGGCTACGAATTCTTGAAGAGGAAATTCTTTTTTATATCATGGATCTCATTAGTGTTATTTCTTTTATGTAACACCTTTATTTATTATAAGGACGAGCAGCAATTTTTTTACGTTAGAAGAATTTTAGGGCTAGGATTATGTGTATCTCGAGCCACTGCGTCGGTACTGAATTTATGCAGCGCATTGCTACTGGTGCCGTTTTGCAAGAAACTAAATCAATTCCTGTACAGGATAATATCGAAATTATGGCCAACATTATTCTTCTATTGGCTGGAGAAAGCGAAAAGCTTTCATATGACTGTGGCGATCACATTGTCCATATTTGCAGTTGTGCATTCTGCTTCTCACTTTGTAAACTTTTGGAATTTCTCAAGAGGATACGACGAAGAATGGACCGAGATTAACTTTGCAAGCTACAAGGATGAGAATCCTCTCTGGCTATTGTTGAGCGTGGCGGGCATAACCGGCGTGTCAATGCTGCTCATAGTCTTGGGTATGAGCGCAACCTCTACTAGGGTTGTTCGACGAAATATGTACAATGTTTTTTGGTATACGCATCAATTGTATCTGATGTTTATGGTGTTACTTATTGTGCACCCATTAAGTGGTGTTCTCAAAGAAGAAGTAGTTAATAGTGAAGCTAGTCTATATTCTAGTCATATTGACCCGTGGTCAAATAACGTTACCTCGGAGGCAATTCCGAAATTTGCGCCCATACAATCGAAGACTTGGTGCTGGATAGCTGTACCATTGACGTGTTACTTAGTCGACATCCTGTGGAGGATATTGACAAGGAACAGGGCGAAGATCGAAATTTTAGAGGTTTACCAGTAA
Protein
MKIKTYLCGGYEFLKRKFFFISWISLVLFLLCNTFIYYKDEQQFFYVRRILGLGLCVSRATASVLNLCSALLLVPFCKKLNQFLYRIISKLWPTLFFYWLEKAKSFHMTVAITLSIFAVVHSASHFVNFWNFSRGYDEEWTEINFASYKDENPLWLLLSVAGITGVSMLLIVLGMSATSTRVVRRNMYNVFWYTHQLYLMFMVLLIVHPLSGVLKEEVVNSEASLYSSHIDPWSNNVTSEAIPKFAPIQSKTWCWIAVPLTCYLVDILWRILTRNRAKIEILEVYQ
Summary
Description
Constitutive NADPH oxidase which generates superoxide intracellularly upon formation of a complex with CYBA/p22phox. Regulates signaling cascades probably through phosphatases inhibition. May function as an oxygen sensor regulating the KCNK3/TASK-1 potassium channel and HIF1A activity. May regulate insulin signaling cascade. May play a role in apoptosis, bone resorption and lipolysaccharide-mediated activation of NFKB (By similarity).
Subunit
Interacts with protein disulfide isomerase (By similarity). Interacts with, relocalizes and stabilizes CYBA/p22phox. Interacts with TLR4.
Interacts with, relocalizes and stabilizes CYBA/p22phox. Interacts with TLR4. Interacts with protein disulfide isomerase (By similarity).
Interacts with, relocalizes and stabilizes CYBA/p22phox. Interacts with TLR4. Interacts with protein disulfide isomerase (By similarity).
Keywords
Alternative splicing
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Membrane
NADP
Nucleus
Oxidoreductase
Polymorphism
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain NADPH oxidase 4
splice variant In isoform 2.
sequence variant In dbSNP:rs317139.
splice variant In isoform 2.
sequence variant In dbSNP:rs317139.
Uniprot
A0A0G3XQZ0
A0A2H1VMB3
A0A2W1BM57
A0A194QRD5
A0A212EKS1
A0A3S2LIV0
+ More
A0A194Q8I0 A0A0L7LKE0 A0A182IN29 A0A1S3GRG5 A0A094L5F6 G3VR45 A0A286XLU3 A0A1Y9GLB7 A0A182QCC0 A0A182WV83 Q7PS77 A0A182LAU5 A0A151MVE9 A0A182UWS6 A0A3Q7TMY3 A0A182F7N9 A0A3Q7UCV7 A0A151MVL8 F6Z703 A0A0N7ZB91 G1LF61 A0A2U9B5E6 D2HYE3 A0A182XXF9 A0A384D7C7 K7GHR7 A0A3Q7XI85 A0A1S3NU63 A0A340XUK4 A0A2Y9G6W1 A0A340XR87 A0A1S3NUL7 A0A2U3WUA1 M3YG53 A0A2U3WUA5 A0A1S3NUM3 A0A2Y9G9C6 A0A2U3X7H2 A0A3Q7QVC9 A0A2Y9K8N1 A0A2Y9KKG5 A0A1Y9H2C9 K7AKP0 A0A2Y9TGR3 A0A2K6FB47 A0A2Y9TAZ9 H2R516 A0A182TKV8 A0A2K5ESJ7 A0A2K6FB25 A0A337SH74 Q9NPH5-6 B7Z523 A0A2K5ESJ6 M3XET3 A0A182P7A5 A0A084VGL7 A0A182JVD9 Q9NPH5 A0A1S3IZ67 Q6V1P7 A0A3F2Z119 L8IHJ1 F1MQX5 A0A2Y9TGQ9 F7CF27 F7CEN8 A0A2K5MB73 A0A2Y9TAZ5 A7E3L3 A0A2Y9TCL3 A0A2K5MB91 I3MB82 A0A2J8S648 Q5R5C5 A0A2I3HSI0 A0A2J8S680 G3THC6 B0WUN3 Q5R5C5-2 H2T6D9 A0A091DFJ6 A0A096MLX2 H2T6E3 A0A2I3LZD7 A0A2K5QKU7 A0A2I3NES7 A0A3Q0GAD5 A0A2K6E3J2 W5P107 S7NES5 A0A2J8MCD2
A0A194Q8I0 A0A0L7LKE0 A0A182IN29 A0A1S3GRG5 A0A094L5F6 G3VR45 A0A286XLU3 A0A1Y9GLB7 A0A182QCC0 A0A182WV83 Q7PS77 A0A182LAU5 A0A151MVE9 A0A182UWS6 A0A3Q7TMY3 A0A182F7N9 A0A3Q7UCV7 A0A151MVL8 F6Z703 A0A0N7ZB91 G1LF61 A0A2U9B5E6 D2HYE3 A0A182XXF9 A0A384D7C7 K7GHR7 A0A3Q7XI85 A0A1S3NU63 A0A340XUK4 A0A2Y9G6W1 A0A340XR87 A0A1S3NUL7 A0A2U3WUA1 M3YG53 A0A2U3WUA5 A0A1S3NUM3 A0A2Y9G9C6 A0A2U3X7H2 A0A3Q7QVC9 A0A2Y9K8N1 A0A2Y9KKG5 A0A1Y9H2C9 K7AKP0 A0A2Y9TGR3 A0A2K6FB47 A0A2Y9TAZ9 H2R516 A0A182TKV8 A0A2K5ESJ7 A0A2K6FB25 A0A337SH74 Q9NPH5-6 B7Z523 A0A2K5ESJ6 M3XET3 A0A182P7A5 A0A084VGL7 A0A182JVD9 Q9NPH5 A0A1S3IZ67 Q6V1P7 A0A3F2Z119 L8IHJ1 F1MQX5 A0A2Y9TGQ9 F7CF27 F7CEN8 A0A2K5MB73 A0A2Y9TAZ5 A7E3L3 A0A2Y9TCL3 A0A2K5MB91 I3MB82 A0A2J8S648 Q5R5C5 A0A2I3HSI0 A0A2J8S680 G3THC6 B0WUN3 Q5R5C5-2 H2T6D9 A0A091DFJ6 A0A096MLX2 H2T6E3 A0A2I3LZD7 A0A2K5QKU7 A0A2I3NES7 A0A3Q0GAD5 A0A2K6E3J2 W5P107 S7NES5 A0A2J8MCD2
EC Number
1.6.3.-
Pubmed
28756777
26354079
22118469
26227816
21709235
21993624
+ More
12364791 14747013 17210077 20966253 22293439 17495919 20010809 25244985 17381049 16136131 17975172 10869423 11376945 11032835 15210697 15721269 14702039 16554811 15489334 15155719 15356101 14966267 15572675 16230378 16179589 16324151 15927447 16019190 23393389 24438588 22751099 19393038 16381931 17612411 21551351 20809919
12364791 14747013 17210077 20966253 22293439 17495919 20010809 25244985 17381049 16136131 17975172 10869423 11376945 11032835 15210697 15721269 14702039 16554811 15489334 15155719 15356101 14966267 15572675 16230378 16179589 16324151 15927447 16019190 23393389 24438588 22751099 19393038 16381931 17612411 21551351 20809919
EMBL
KP666171
AKM16733.1
ODYU01003336
SOQ41957.1
KZ149998
PZC75371.1
+ More
KQ461175 KPJ07904.1 AGBW02014216 OWR42086.1 RSAL01000003 RVE54790.1 KQ459302 KPJ01704.1 JTDY01000777 KOB75917.1 KL359819 KFZ64677.1 AEFK01119641 AEFK01119642 AEFK01119643 AEFK01119644 AEFK01119645 AEFK01119646 AEFK01119647 AEFK01119648 AEFK01119649 AEFK01119650 AAKN02037747 AAKN02037748 AAKN02037749 AAKN02037750 AAKN02037751 AAKN02037752 APCN01003125 AXCN02000040 AAAB01008844 EAA06046.6 AKHW03004889 KYO28533.1 KYO28534.1 GDRN01087748 JAI60992.1 ACTA01035104 ACTA01043103 ACTA01051103 ACTA01059103 ACTA01067103 ACTA01075103 ACTA01083103 ACTA01091103 ACTA01099103 ACTA01107103 CP026245 AWO99134.1 GL193693 EFB16026.1 AGCU01053606 AGCU01053607 AGCU01053608 AGCU01053609 AGCU01053610 AGCU01053611 AGCU01053612 AGCU01053613 AEYP01058592 AEYP01058593 AEYP01058594 AACZ04016362 AACZ04016363 GABF01003754 NBAG03000261 JAA18391.1 PNI57173.1 GABF01003753 JAA18392.1 PNI57175.1 AANG04002890 AF261943 AF254621 AB041035 AY288918 AJ704725 AJ704726 AJ704727 AJ704728 AJ704729 AK291830 AK298323 AP003400 AP001815 AP002404 BC040105 BC051371 AK298328 BAH12759.1 ATLV01012999 KE524833 KFB37111.1 AY354499 AAQ72353.1 JH881171 ELR56020.1 BR000275 FAA00342.1 AGTP01101097 AGTP01101098 AGTP01101099 AGTP01101100 AGTP01101101 AGTP01101102 AGTP01101103 AGTP01101104 NDHI03003605 PNJ16250.1 CR857676 CR858307 CR860937 ADFV01110741 ADFV01110742 ADFV01110743 ADFV01110744 ADFV01110745 ADFV01110746 ADFV01110747 PNJ16254.1 DS232107 EDS35018.1 KN122620 KFO29238.1 AHZZ02007286 AHZZ02007287 AMGL01060390 AMGL01060391 AMGL01060392 AMGL01060393 AMGL01060394 KE164192 EPQ15849.1 PNI57177.1
KQ461175 KPJ07904.1 AGBW02014216 OWR42086.1 RSAL01000003 RVE54790.1 KQ459302 KPJ01704.1 JTDY01000777 KOB75917.1 KL359819 KFZ64677.1 AEFK01119641 AEFK01119642 AEFK01119643 AEFK01119644 AEFK01119645 AEFK01119646 AEFK01119647 AEFK01119648 AEFK01119649 AEFK01119650 AAKN02037747 AAKN02037748 AAKN02037749 AAKN02037750 AAKN02037751 AAKN02037752 APCN01003125 AXCN02000040 AAAB01008844 EAA06046.6 AKHW03004889 KYO28533.1 KYO28534.1 GDRN01087748 JAI60992.1 ACTA01035104 ACTA01043103 ACTA01051103 ACTA01059103 ACTA01067103 ACTA01075103 ACTA01083103 ACTA01091103 ACTA01099103 ACTA01107103 CP026245 AWO99134.1 GL193693 EFB16026.1 AGCU01053606 AGCU01053607 AGCU01053608 AGCU01053609 AGCU01053610 AGCU01053611 AGCU01053612 AGCU01053613 AEYP01058592 AEYP01058593 AEYP01058594 AACZ04016362 AACZ04016363 GABF01003754 NBAG03000261 JAA18391.1 PNI57173.1 GABF01003753 JAA18392.1 PNI57175.1 AANG04002890 AF261943 AF254621 AB041035 AY288918 AJ704725 AJ704726 AJ704727 AJ704728 AJ704729 AK291830 AK298323 AP003400 AP001815 AP002404 BC040105 BC051371 AK298328 BAH12759.1 ATLV01012999 KE524833 KFB37111.1 AY354499 AAQ72353.1 JH881171 ELR56020.1 BR000275 FAA00342.1 AGTP01101097 AGTP01101098 AGTP01101099 AGTP01101100 AGTP01101101 AGTP01101102 AGTP01101103 AGTP01101104 NDHI03003605 PNJ16250.1 CR857676 CR858307 CR860937 ADFV01110741 ADFV01110742 ADFV01110743 ADFV01110744 ADFV01110745 ADFV01110746 ADFV01110747 PNJ16254.1 DS232107 EDS35018.1 KN122620 KFO29238.1 AHZZ02007286 AHZZ02007287 AMGL01060390 AMGL01060391 AMGL01060392 AMGL01060393 AMGL01060394 KE164192 EPQ15849.1 PNI57177.1
Proteomes
UP000053240
UP000007151
UP000283053
UP000053268
UP000037510
UP000075880
+ More
UP000081671 UP000007648 UP000005447 UP000075840 UP000075886 UP000076407 UP000007062 UP000075882 UP000050525 UP000075903 UP000286640 UP000069272 UP000002280 UP000008912 UP000246464 UP000076408 UP000261680 UP000007267 UP000286642 UP000087266 UP000265300 UP000248481 UP000245340 UP000000715 UP000245341 UP000286641 UP000248482 UP000075884 UP000002277 UP000248484 UP000233160 UP000075902 UP000233020 UP000011712 UP000005640 UP000075885 UP000030765 UP000075881 UP000085678 UP000075920 UP000009136 UP000008225 UP000233060 UP000005215 UP000001595 UP000001073 UP000007646 UP000002320 UP000005226 UP000028990 UP000028761 UP000233040 UP000189705 UP000233120 UP000002356
UP000081671 UP000007648 UP000005447 UP000075840 UP000075886 UP000076407 UP000007062 UP000075882 UP000050525 UP000075903 UP000286640 UP000069272 UP000002280 UP000008912 UP000246464 UP000076408 UP000261680 UP000007267 UP000286642 UP000087266 UP000265300 UP000248481 UP000245340 UP000000715 UP000245341 UP000286641 UP000248482 UP000075884 UP000002277 UP000248484 UP000233160 UP000075902 UP000233020 UP000011712 UP000005640 UP000075885 UP000030765 UP000075881 UP000085678 UP000075920 UP000009136 UP000008225 UP000233060 UP000005215 UP000001595 UP000001073 UP000007646 UP000002320 UP000005226 UP000028990 UP000028761 UP000233040 UP000189705 UP000233120 UP000002356
Interpro
Gene 3D
ProteinModelPortal
A0A0G3XQZ0
A0A2H1VMB3
A0A2W1BM57
A0A194QRD5
A0A212EKS1
A0A3S2LIV0
+ More
A0A194Q8I0 A0A0L7LKE0 A0A182IN29 A0A1S3GRG5 A0A094L5F6 G3VR45 A0A286XLU3 A0A1Y9GLB7 A0A182QCC0 A0A182WV83 Q7PS77 A0A182LAU5 A0A151MVE9 A0A182UWS6 A0A3Q7TMY3 A0A182F7N9 A0A3Q7UCV7 A0A151MVL8 F6Z703 A0A0N7ZB91 G1LF61 A0A2U9B5E6 D2HYE3 A0A182XXF9 A0A384D7C7 K7GHR7 A0A3Q7XI85 A0A1S3NU63 A0A340XUK4 A0A2Y9G6W1 A0A340XR87 A0A1S3NUL7 A0A2U3WUA1 M3YG53 A0A2U3WUA5 A0A1S3NUM3 A0A2Y9G9C6 A0A2U3X7H2 A0A3Q7QVC9 A0A2Y9K8N1 A0A2Y9KKG5 A0A1Y9H2C9 K7AKP0 A0A2Y9TGR3 A0A2K6FB47 A0A2Y9TAZ9 H2R516 A0A182TKV8 A0A2K5ESJ7 A0A2K6FB25 A0A337SH74 Q9NPH5-6 B7Z523 A0A2K5ESJ6 M3XET3 A0A182P7A5 A0A084VGL7 A0A182JVD9 Q9NPH5 A0A1S3IZ67 Q6V1P7 A0A3F2Z119 L8IHJ1 F1MQX5 A0A2Y9TGQ9 F7CF27 F7CEN8 A0A2K5MB73 A0A2Y9TAZ5 A7E3L3 A0A2Y9TCL3 A0A2K5MB91 I3MB82 A0A2J8S648 Q5R5C5 A0A2I3HSI0 A0A2J8S680 G3THC6 B0WUN3 Q5R5C5-2 H2T6D9 A0A091DFJ6 A0A096MLX2 H2T6E3 A0A2I3LZD7 A0A2K5QKU7 A0A2I3NES7 A0A3Q0GAD5 A0A2K6E3J2 W5P107 S7NES5 A0A2J8MCD2
A0A194Q8I0 A0A0L7LKE0 A0A182IN29 A0A1S3GRG5 A0A094L5F6 G3VR45 A0A286XLU3 A0A1Y9GLB7 A0A182QCC0 A0A182WV83 Q7PS77 A0A182LAU5 A0A151MVE9 A0A182UWS6 A0A3Q7TMY3 A0A182F7N9 A0A3Q7UCV7 A0A151MVL8 F6Z703 A0A0N7ZB91 G1LF61 A0A2U9B5E6 D2HYE3 A0A182XXF9 A0A384D7C7 K7GHR7 A0A3Q7XI85 A0A1S3NU63 A0A340XUK4 A0A2Y9G6W1 A0A340XR87 A0A1S3NUL7 A0A2U3WUA1 M3YG53 A0A2U3WUA5 A0A1S3NUM3 A0A2Y9G9C6 A0A2U3X7H2 A0A3Q7QVC9 A0A2Y9K8N1 A0A2Y9KKG5 A0A1Y9H2C9 K7AKP0 A0A2Y9TGR3 A0A2K6FB47 A0A2Y9TAZ9 H2R516 A0A182TKV8 A0A2K5ESJ7 A0A2K6FB25 A0A337SH74 Q9NPH5-6 B7Z523 A0A2K5ESJ6 M3XET3 A0A182P7A5 A0A084VGL7 A0A182JVD9 Q9NPH5 A0A1S3IZ67 Q6V1P7 A0A3F2Z119 L8IHJ1 F1MQX5 A0A2Y9TGQ9 F7CF27 F7CEN8 A0A2K5MB73 A0A2Y9TAZ5 A7E3L3 A0A2Y9TCL3 A0A2K5MB91 I3MB82 A0A2J8S648 Q5R5C5 A0A2I3HSI0 A0A2J8S680 G3THC6 B0WUN3 Q5R5C5-2 H2T6D9 A0A091DFJ6 A0A096MLX2 H2T6E3 A0A2I3LZD7 A0A2K5QKU7 A0A2I3NES7 A0A3Q0GAD5 A0A2K6E3J2 W5P107 S7NES5 A0A2J8MCD2
PDB
5O0T
E-value=4.09008e-05,
Score=110
Ontologies
PATHWAY
GO
GO:0016491
GO:0016021
GO:0050667
GO:0005634
GO:0010467
GO:0072341
GO:0006801
GO:0003015
GO:0061098
GO:0042554
GO:0045453
GO:0051897
GO:0016175
GO:0055007
GO:0070374
GO:2000573
GO:0007569
GO:0000902
GO:0048471
GO:0071333
GO:1990782
GO:0008285
GO:0043406
GO:0071944
GO:0006952
GO:0005886
GO:0055114
GO:0043020
GO:0005925
GO:0014911
GO:0034599
GO:0071320
GO:0001725
GO:0097038
GO:0005739
GO:0001666
GO:0016174
GO:0005730
GO:0016324
GO:0005789
GO:0019826
GO:0000166
GO:2000379
GO:0020037
GO:0071480
GO:0051496
GO:0050660
GO:0071560
GO:0006954
GO:0072593
GO:1903409
GO:0009055
GO:0043065
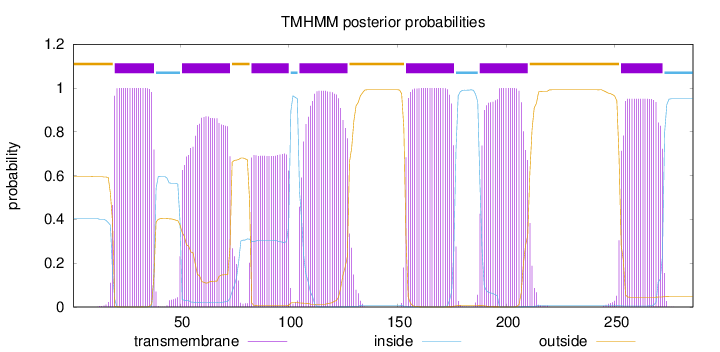
Topology
Subcellular location
Endoplasmic reticulum membrane
Cell membrane
Cell junction
Focal adhesion
Nucleus
Nucleolus
Cell membrane
Cell junction
Focal adhesion
Nucleus
Nucleolus
Length:
286
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
136.24703
Exp number, first 60 AAs:
27.19228
Total prob of N-in:
0.40431
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 38
inside
39 - 50
TMhelix
51 - 73
outside
74 - 82
TMhelix
83 - 100
inside
101 - 104
TMhelix
105 - 127
outside
128 - 153
TMhelix
154 - 176
inside
177 - 187
TMhelix
188 - 210
outside
211 - 252
TMhelix
253 - 272
inside
273 - 286
Population Genetic Test Statistics
Pi
195.327584
Theta
177.927721
Tajima's D
0.332707
CLR
0.180246
CSRT
0.464476776161192
Interpretation
Uncertain
