Pre Gene Modal
BGIBMGA002933
Annotation
Vesicle_transport_protein_SEC20_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.47
Sequence
CDS
ATGTTCTGCTTTGCACCAAATATTCTCAAATATGAAGTAAAATACACCACAATGGCCCTCAAAATGGATTTGAGTCCAACGGACCCATATCATCAATCGTTGTTTGCAAAATTAGAAAATCCTGATGTGGTCTATCAGAATTTCGTGAAGAGATTGGCTAAGTATCATTTTCAAGTCAAAGCAATTATCCAGGATATCCTGGAATGCAGGGAGTCGATGGAGAAATTAAATGAGTTAAATGAAAAAGGAAGGGCAAAAATAACGGAAGTACGAGAGGAATTAGAAAATTTATACCTGTATGGTAAAGAAACAGGGGATACAAAATATGATACTGAATTAGAATCACAGAGGCACCTTTTAGCTTGTCTCCTTAAGGAATTTAAAGATGCCAATATAAATAGTATGTTTGCTATTGAAAAGGCACAAAGAGAAGATTTATTAAAATGTAGTGATAATGAGAAGTCATCAGTCCGACAAAGAAAGAAAAAGATGGATAAGGATGGATTGGTCAAAATGTCATCAGGTGTGACAGAGCAATTACTTTCCGTAAGCAGGCAGCTAGCGGACACCACACAAAAGAGTCAAAATACTTTGGATAATTTGGTTTCATCTAGTTCAACAGTGCATGGAACACAATCGGAATTAGAGAATACAGCAGGAACGATTACGCAATCGAGCAAACTACTGAAGAAATACGGACGTAGAGAATTTACGGACAAAGTGATCATGTTTTTTGCATTTTTATTCTTTTTGGCGGTGTGCCTGTACATTGTACAGAAGAGGCTTTTTTAA
Protein
MFCFAPNILKYEVKYTTMALKMDLSPTDPYHQSLFAKLENPDVVYQNFVKRLAKYHFQVKAIIQDILECRESMEKLNELNEKGRAKITEVREELENLYLYGKETGDTKYDTELESQRHLLACLLKEFKDANINSMFAIEKAQREDLLKCSDNEKSSVRQRKKKMDKDGLVKMSSGVTEQLLSVSRQLADTTQKSQNTLDNLVSSSSTVHGTQSELENTAGTITQSSKLLKKYGRREFTDKVIMFFAFLFFLAVCLYIVQKRLF
Summary
Uniprot
H9J098
A0A3S2P891
A0A1E1W2Z1
A0A2A4IYW2
A0A2W1BJZ9
A0A194QQY1
+ More
A0A194QE17 A0A1E1WU75 A0A2H1WK56 A0A212EKS9 S4PWU0 A0A1W4X0D8 D6X2A5 R4V329 A0A1B0D024 A0A1B6L192 A0A1L8DUH8 A0A1B6JI59 A0A1A9Y8L5 A0A1B0BRF4 A0A1A9VLK3 A0A034WNH8 B4KBE7 A0A0A9YT10 A0A0K8VCI3 B4NAE8 A0A1A9ZN44 B4MB72 A0A146LD63 B4JSC8 A0A1B6GP19 T1PJ33 A0A0A1WFE7 A0A1Y1L2Q8 A0A0L0CRH7 B3LWD0 B3NYH8 A0A1W4UXA2 U5ETS1 A0A1I8Q799 B4I483 B4QXL0 W8CCR3 A0A0K8TN84 N6U8K3 Q9VNI9 B4PVI9 B4G5A1 Q299J7 A0A1A9X2K5 A0A3B0K4D2 A0A2J7Q7N8 A0A0M5J1Z4 A0A1J1HS09 A0A1S4FDE9 Q176G1 B0WE04 J3JVG8 A0A336KTI9 A0A336NA81 A0A1Q3FGU1 A0A023EKQ3 A0A1Q3FGJ0 A0A1Q3FGI8 A0A2S2PXJ8 A0A0P4VYS2 R4FN77 A0A0L7RBK0 A0A0J7KJS2 A0A067R6L2 A0A026WP85 J9K6I3 W5JCG7 E2BB72 A0A2M4BZD2 A0A2M4CZ72 A0A023F9C4 A0A2M4CZ22 A0A151WM64 A0A2P8YX98 A0A182FJ32 K7JA57 A0A2M4ANJ1 A0A0C9QGT6 A0A232EEM1 A0A088ABZ2 A0A2A3E7U5 A0A182Q2G9 A0A195E5K3 A0A2M3Z8Y4 A0A182SN51 A0A182KH60 A0A195CMD1 A0A182N0B7 A0A182RQ74 A0A182PT73 A0A182MBK4 F4WP26
A0A194QE17 A0A1E1WU75 A0A2H1WK56 A0A212EKS9 S4PWU0 A0A1W4X0D8 D6X2A5 R4V329 A0A1B0D024 A0A1B6L192 A0A1L8DUH8 A0A1B6JI59 A0A1A9Y8L5 A0A1B0BRF4 A0A1A9VLK3 A0A034WNH8 B4KBE7 A0A0A9YT10 A0A0K8VCI3 B4NAE8 A0A1A9ZN44 B4MB72 A0A146LD63 B4JSC8 A0A1B6GP19 T1PJ33 A0A0A1WFE7 A0A1Y1L2Q8 A0A0L0CRH7 B3LWD0 B3NYH8 A0A1W4UXA2 U5ETS1 A0A1I8Q799 B4I483 B4QXL0 W8CCR3 A0A0K8TN84 N6U8K3 Q9VNI9 B4PVI9 B4G5A1 Q299J7 A0A1A9X2K5 A0A3B0K4D2 A0A2J7Q7N8 A0A0M5J1Z4 A0A1J1HS09 A0A1S4FDE9 Q176G1 B0WE04 J3JVG8 A0A336KTI9 A0A336NA81 A0A1Q3FGU1 A0A023EKQ3 A0A1Q3FGJ0 A0A1Q3FGI8 A0A2S2PXJ8 A0A0P4VYS2 R4FN77 A0A0L7RBK0 A0A0J7KJS2 A0A067R6L2 A0A026WP85 J9K6I3 W5JCG7 E2BB72 A0A2M4BZD2 A0A2M4CZ72 A0A023F9C4 A0A2M4CZ22 A0A151WM64 A0A2P8YX98 A0A182FJ32 K7JA57 A0A2M4ANJ1 A0A0C9QGT6 A0A232EEM1 A0A088ABZ2 A0A2A3E7U5 A0A182Q2G9 A0A195E5K3 A0A2M3Z8Y4 A0A182SN51 A0A182KH60 A0A195CMD1 A0A182N0B7 A0A182RQ74 A0A182PT73 A0A182MBK4 F4WP26
Pubmed
19121390
28756777
26354079
22118469
23622113
18362917
+ More
19820115 25348373 17994087 25401762 26823975 25315136 25830018 28004739 26108605 24495485 26369729 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 17510324 22516182 24945155 26483478 27129103 24845553 24508170 30249741 20920257 23761445 20798317 25474469 29403074 20075255 28648823 21719571
19820115 25348373 17994087 25401762 26823975 25315136 25830018 28004739 26108605 24495485 26369729 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 17510324 22516182 24945155 26483478 27129103 24845553 24508170 30249741 20920257 23761445 20798317 25474469 29403074 20075255 28648823 21719571
EMBL
BABH01019173
RSAL01000003
RVE54792.1
GDQN01009714
JAT81340.1
NWSH01004649
+ More
PCG64819.1 KZ149998 PZC75369.1 KQ461175 KPJ07902.1 KQ459302 KPJ01706.1 GDQN01000455 JAT90599.1 ODYU01009104 SOQ53282.1 AGBW02014216 OWR42089.1 GAIX01006203 JAA86357.1 KQ971371 EFA09888.1 KC632374 AGM32188.1 AJVK01021187 GEBQ01022498 JAT17479.1 GFDF01004004 JAV10080.1 GECU01008720 JAS98986.1 JXJN01019080 GAKP01003040 GAKP01003039 JAC55912.1 CH933806 EDW16875.2 GBHO01008265 GBRD01010631 GDHC01020123 GDHC01005219 JAG35339.1 JAG55193.1 JAP98505.1 JAQ13410.1 GDHF01015650 JAI36664.1 CH964232 EDW80762.1 CH940656 EDW58343.1 GDHC01012431 JAQ06198.1 CH916373 EDV94668.1 GECZ01005610 JAS64159.1 KA648694 AFP63323.1 GBXI01017092 JAC97199.1 GEZM01066326 JAV67962.1 JRES01000007 KNC34918.1 CH902617 EDV43763.1 CH954181 EDV47959.1 GANO01001777 JAB58094.1 CH480821 EDW55026.1 CM000364 EDX11796.1 GAMC01003246 JAC03310.1 GDAI01001761 JAI15842.1 APGK01044468 KB741026 KB632309 ENN74932.1 ERL92095.1 AE014297 BT030855 AAF51945.1 ABV82237.1 CM000160 EDW97798.1 CH479179 EDW24767.1 CM000070 EAL27706.1 OUUW01000007 SPP82850.1 NEVH01017441 PNF24603.1 CP012526 ALC46074.1 CVRI01000010 CRK88985.1 CH477387 EAT42055.1 DS231903 EDS45134.1 BT127236 AEE62198.1 UFQS01000878 UFQT01000878 SSX07452.1 SSX27792.1 UFQT01004575 SSX35718.1 GFDL01008258 JAV26787.1 JXUM01141297 GAPW01003858 KQ569220 JAC09740.1 KXJ68713.1 GFDL01008379 JAV26666.1 GFDL01008433 JAV26612.1 GGMS01000978 MBY70181.1 GDKW01000828 JAI55767.1 ACPB03011791 GAHY01000668 JAA76842.1 KQ414617 KOC68307.1 LBMM01006354 KMQ90673.1 KK852710 KDR17970.1 KK107139 QOIP01000003 EZA57773.1 RLU24737.1 ABLF02021570 ADMH02001819 ETN61023.1 GL446946 EFN87045.1 GGFJ01008957 MBW58098.1 GGFL01006401 MBW70579.1 GBBI01001113 JAC17599.1 GGFL01006402 MBW70580.1 KQ982944 KYQ48908.1 PYGN01000304 PSN48876.1 AAZX01001625 GGFK01008981 MBW42302.1 GBYB01002699 JAG72466.1 NNAY01005274 OXU16801.1 KZ288338 PBC27833.1 AXCN02001229 KQ979608 KYN20366.1 GGFM01004199 MBW24950.1 KQ977574 KYN01886.1 AXCM01000049 GL888243 EGI63957.1
PCG64819.1 KZ149998 PZC75369.1 KQ461175 KPJ07902.1 KQ459302 KPJ01706.1 GDQN01000455 JAT90599.1 ODYU01009104 SOQ53282.1 AGBW02014216 OWR42089.1 GAIX01006203 JAA86357.1 KQ971371 EFA09888.1 KC632374 AGM32188.1 AJVK01021187 GEBQ01022498 JAT17479.1 GFDF01004004 JAV10080.1 GECU01008720 JAS98986.1 JXJN01019080 GAKP01003040 GAKP01003039 JAC55912.1 CH933806 EDW16875.2 GBHO01008265 GBRD01010631 GDHC01020123 GDHC01005219 JAG35339.1 JAG55193.1 JAP98505.1 JAQ13410.1 GDHF01015650 JAI36664.1 CH964232 EDW80762.1 CH940656 EDW58343.1 GDHC01012431 JAQ06198.1 CH916373 EDV94668.1 GECZ01005610 JAS64159.1 KA648694 AFP63323.1 GBXI01017092 JAC97199.1 GEZM01066326 JAV67962.1 JRES01000007 KNC34918.1 CH902617 EDV43763.1 CH954181 EDV47959.1 GANO01001777 JAB58094.1 CH480821 EDW55026.1 CM000364 EDX11796.1 GAMC01003246 JAC03310.1 GDAI01001761 JAI15842.1 APGK01044468 KB741026 KB632309 ENN74932.1 ERL92095.1 AE014297 BT030855 AAF51945.1 ABV82237.1 CM000160 EDW97798.1 CH479179 EDW24767.1 CM000070 EAL27706.1 OUUW01000007 SPP82850.1 NEVH01017441 PNF24603.1 CP012526 ALC46074.1 CVRI01000010 CRK88985.1 CH477387 EAT42055.1 DS231903 EDS45134.1 BT127236 AEE62198.1 UFQS01000878 UFQT01000878 SSX07452.1 SSX27792.1 UFQT01004575 SSX35718.1 GFDL01008258 JAV26787.1 JXUM01141297 GAPW01003858 KQ569220 JAC09740.1 KXJ68713.1 GFDL01008379 JAV26666.1 GFDL01008433 JAV26612.1 GGMS01000978 MBY70181.1 GDKW01000828 JAI55767.1 ACPB03011791 GAHY01000668 JAA76842.1 KQ414617 KOC68307.1 LBMM01006354 KMQ90673.1 KK852710 KDR17970.1 KK107139 QOIP01000003 EZA57773.1 RLU24737.1 ABLF02021570 ADMH02001819 ETN61023.1 GL446946 EFN87045.1 GGFJ01008957 MBW58098.1 GGFL01006401 MBW70579.1 GBBI01001113 JAC17599.1 GGFL01006402 MBW70580.1 KQ982944 KYQ48908.1 PYGN01000304 PSN48876.1 AAZX01001625 GGFK01008981 MBW42302.1 GBYB01002699 JAG72466.1 NNAY01005274 OXU16801.1 KZ288338 PBC27833.1 AXCN02001229 KQ979608 KYN20366.1 GGFM01004199 MBW24950.1 KQ977574 KYN01886.1 AXCM01000049 GL888243 EGI63957.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000192223 UP000007266 UP000092462 UP000092443 UP000092460 UP000078200 UP000009192 UP000007798 UP000092445 UP000008792 UP000001070 UP000095301 UP000037069 UP000007801 UP000008711 UP000192221 UP000095300 UP000001292 UP000000304 UP000019118 UP000030742 UP000000803 UP000002282 UP000008744 UP000001819 UP000091820 UP000268350 UP000235965 UP000092553 UP000183832 UP000008820 UP000002320 UP000069940 UP000249989 UP000015103 UP000053825 UP000036403 UP000027135 UP000053097 UP000279307 UP000007819 UP000000673 UP000008237 UP000075809 UP000245037 UP000069272 UP000002358 UP000215335 UP000005203 UP000242457 UP000075886 UP000078492 UP000075901 UP000075881 UP000078542 UP000075884 UP000075900 UP000075885 UP000075883 UP000007755
UP000192223 UP000007266 UP000092462 UP000092443 UP000092460 UP000078200 UP000009192 UP000007798 UP000092445 UP000008792 UP000001070 UP000095301 UP000037069 UP000007801 UP000008711 UP000192221 UP000095300 UP000001292 UP000000304 UP000019118 UP000030742 UP000000803 UP000002282 UP000008744 UP000001819 UP000091820 UP000268350 UP000235965 UP000092553 UP000183832 UP000008820 UP000002320 UP000069940 UP000249989 UP000015103 UP000053825 UP000036403 UP000027135 UP000053097 UP000279307 UP000007819 UP000000673 UP000008237 UP000075809 UP000245037 UP000069272 UP000002358 UP000215335 UP000005203 UP000242457 UP000075886 UP000078492 UP000075901 UP000075881 UP000078542 UP000075884 UP000075900 UP000075885 UP000075883 UP000007755
Pfam
PF03908 Sec20
ProteinModelPortal
H9J098
A0A3S2P891
A0A1E1W2Z1
A0A2A4IYW2
A0A2W1BJZ9
A0A194QQY1
+ More
A0A194QE17 A0A1E1WU75 A0A2H1WK56 A0A212EKS9 S4PWU0 A0A1W4X0D8 D6X2A5 R4V329 A0A1B0D024 A0A1B6L192 A0A1L8DUH8 A0A1B6JI59 A0A1A9Y8L5 A0A1B0BRF4 A0A1A9VLK3 A0A034WNH8 B4KBE7 A0A0A9YT10 A0A0K8VCI3 B4NAE8 A0A1A9ZN44 B4MB72 A0A146LD63 B4JSC8 A0A1B6GP19 T1PJ33 A0A0A1WFE7 A0A1Y1L2Q8 A0A0L0CRH7 B3LWD0 B3NYH8 A0A1W4UXA2 U5ETS1 A0A1I8Q799 B4I483 B4QXL0 W8CCR3 A0A0K8TN84 N6U8K3 Q9VNI9 B4PVI9 B4G5A1 Q299J7 A0A1A9X2K5 A0A3B0K4D2 A0A2J7Q7N8 A0A0M5J1Z4 A0A1J1HS09 A0A1S4FDE9 Q176G1 B0WE04 J3JVG8 A0A336KTI9 A0A336NA81 A0A1Q3FGU1 A0A023EKQ3 A0A1Q3FGJ0 A0A1Q3FGI8 A0A2S2PXJ8 A0A0P4VYS2 R4FN77 A0A0L7RBK0 A0A0J7KJS2 A0A067R6L2 A0A026WP85 J9K6I3 W5JCG7 E2BB72 A0A2M4BZD2 A0A2M4CZ72 A0A023F9C4 A0A2M4CZ22 A0A151WM64 A0A2P8YX98 A0A182FJ32 K7JA57 A0A2M4ANJ1 A0A0C9QGT6 A0A232EEM1 A0A088ABZ2 A0A2A3E7U5 A0A182Q2G9 A0A195E5K3 A0A2M3Z8Y4 A0A182SN51 A0A182KH60 A0A195CMD1 A0A182N0B7 A0A182RQ74 A0A182PT73 A0A182MBK4 F4WP26
A0A194QE17 A0A1E1WU75 A0A2H1WK56 A0A212EKS9 S4PWU0 A0A1W4X0D8 D6X2A5 R4V329 A0A1B0D024 A0A1B6L192 A0A1L8DUH8 A0A1B6JI59 A0A1A9Y8L5 A0A1B0BRF4 A0A1A9VLK3 A0A034WNH8 B4KBE7 A0A0A9YT10 A0A0K8VCI3 B4NAE8 A0A1A9ZN44 B4MB72 A0A146LD63 B4JSC8 A0A1B6GP19 T1PJ33 A0A0A1WFE7 A0A1Y1L2Q8 A0A0L0CRH7 B3LWD0 B3NYH8 A0A1W4UXA2 U5ETS1 A0A1I8Q799 B4I483 B4QXL0 W8CCR3 A0A0K8TN84 N6U8K3 Q9VNI9 B4PVI9 B4G5A1 Q299J7 A0A1A9X2K5 A0A3B0K4D2 A0A2J7Q7N8 A0A0M5J1Z4 A0A1J1HS09 A0A1S4FDE9 Q176G1 B0WE04 J3JVG8 A0A336KTI9 A0A336NA81 A0A1Q3FGU1 A0A023EKQ3 A0A1Q3FGJ0 A0A1Q3FGI8 A0A2S2PXJ8 A0A0P4VYS2 R4FN77 A0A0L7RBK0 A0A0J7KJS2 A0A067R6L2 A0A026WP85 J9K6I3 W5JCG7 E2BB72 A0A2M4BZD2 A0A2M4CZ72 A0A023F9C4 A0A2M4CZ22 A0A151WM64 A0A2P8YX98 A0A182FJ32 K7JA57 A0A2M4ANJ1 A0A0C9QGT6 A0A232EEM1 A0A088ABZ2 A0A2A3E7U5 A0A182Q2G9 A0A195E5K3 A0A2M3Z8Y4 A0A182SN51 A0A182KH60 A0A195CMD1 A0A182N0B7 A0A182RQ74 A0A182PT73 A0A182MBK4 F4WP26
Ontologies
GO
PANTHER
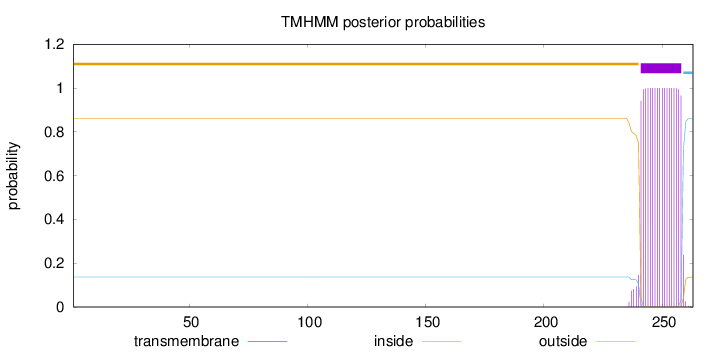
Topology
Length:
263
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.58133
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13752
outside
1 - 240
TMhelix
241 - 258
inside
259 - 263
Population Genetic Test Statistics
Pi
36.592186
Theta
33.703487
Tajima's D
0.165121
CLR
0.249748
CSRT
0.417779111044448
Interpretation
Uncertain
