Gene
KWMTBOMO05882
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.023 Nuclear Reliability : 2.362
Sequence
CDS
ATGTATCTAGCCTATCGTCATGTATGGTTGCGAAACGTGGACCCTGAAAGAGGACACTACAAGACGCCTACAGGCATTCGAGATGTGGTGCTACCGTCGCATGTTACGCAGTTTAGTTGGACGCAGAGGGTCAGGAACGAAACCGTGCTTCAGCGCGTTCATATGTCCCGGAAAATGTTGCCTGCTATCAAAAAGCGCAAGATAGAGTATCTCGGCCACGTGCTCAGGAACGATCGATACATGCTGCTGCAGCTGATAATTATGGGCAAAGTGGACGGAAAGAGACGTGCTGGGCGAAGAAAAAAGTCATGGCTCCGGAATATCCGGGAGTGGACCAGTATTGCCTCCGTCGAACAGCTGTTCCGGCTGGCTGCTGACAGAAAGGAATACAGAAAACTAACGGCCAACCTTCAGGATTGA
Protein
MYLAYRHVWLRNVDPERGHYKTPTGIRDVVLPSHVTQFSWTQRVRNETVLQRVHMSRKMLPAIKKRKIEYLGHVLRNDRYMLLQLIIMGKVDGKRRAGRRKKSWLRNIREWTSIASVEQLFRLAADRKEYRKLTANLQD
Summary
Uniprot
D5LB38
A0A2A4J795
A0A151IIH7
A0A146L0Z1
A0A2H1V1U0
A0A0K8T3U8
+ More
A0A2S2QJK3 A0A0A9XCF5 A0A0J7KRQ5 A0A0J7KZE8 A0A2A4JBK9 A0A2S2QAF2 A0A2S2PW26 A0A2W1BSK1 A0A0J7KK86 W4Y921 X1D4L4 A0A3S1AZH6 A0A3S0ZE77 J9LIJ7 A0A2S2P9Y7 A0A2S2P7C6 A0A3S1AY56 A0A067QXS5 A0A0B7BUS3 A0A0B7BSZ9 A0A0B7BT80 A0A0B7BUR9 A0A3S1A2L0 A0A3S1CD12 A0A067RNC1 A0A067QUH9 A0A1W7RAL1 A0A3S0ZU31 A0A3S1HJZ0 A0A2J7PZP5 A0A067QJX4
A0A2S2QJK3 A0A0A9XCF5 A0A0J7KRQ5 A0A0J7KZE8 A0A2A4JBK9 A0A2S2QAF2 A0A2S2PW26 A0A2W1BSK1 A0A0J7KK86 W4Y921 X1D4L4 A0A3S1AZH6 A0A3S0ZE77 J9LIJ7 A0A2S2P9Y7 A0A2S2P7C6 A0A3S1AY56 A0A067QXS5 A0A0B7BUS3 A0A0B7BSZ9 A0A0B7BT80 A0A0B7BUR9 A0A3S1A2L0 A0A3S1CD12 A0A067RNC1 A0A067QUH9 A0A1W7RAL1 A0A3S0ZU31 A0A3S1HJZ0 A0A2J7PZP5 A0A067QJX4
EMBL
GU815089
ADF18552.1
NWSH01002736
PCG67606.1
KQ977522
KYN02133.1
+ More
GDHC01017214 JAQ01415.1 ODYU01000306 SOQ34818.1 GBRD01005612 JAG60209.1 GGMS01008711 MBY77914.1 GBHO01028834 JAG14770.1 LBMM01003995 KMQ92919.1 LBMM01001656 KMQ95932.1 NWSH01001998 PCG69477.1 GGMS01004979 MBY74182.1 GGMS01000367 MBY69570.1 KZ149912 PZC78062.1 LBMM01006413 KMQ90631.1 AAGJ04161766 BART01031562 GAH15691.1 RQTK01000685 RUS76136.1 RQTK01000625 RUS76894.1 ABLF02012409 ABLF02012412 GGMR01013640 MBY26259.1 GGMR01012724 MBY25343.1 RQTK01001448 RUS70261.1 KK853131 KDR10935.1 HACG01049256 CEK96121.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 RQTK01000357 RUS81065.1 RQTK01000058 RUS89376.1 KK852404 KDR24543.1 KK852941 KDR13536.1 GFAH01000222 JAV48167.1 RQTK01000752 RUS75316.1 RQTK01000360 RUS81011.1 NEVH01020333 PNF21801.1 KK853249 KDR09341.1
GDHC01017214 JAQ01415.1 ODYU01000306 SOQ34818.1 GBRD01005612 JAG60209.1 GGMS01008711 MBY77914.1 GBHO01028834 JAG14770.1 LBMM01003995 KMQ92919.1 LBMM01001656 KMQ95932.1 NWSH01001998 PCG69477.1 GGMS01004979 MBY74182.1 GGMS01000367 MBY69570.1 KZ149912 PZC78062.1 LBMM01006413 KMQ90631.1 AAGJ04161766 BART01031562 GAH15691.1 RQTK01000685 RUS76136.1 RQTK01000625 RUS76894.1 ABLF02012409 ABLF02012412 GGMR01013640 MBY26259.1 GGMR01012724 MBY25343.1 RQTK01001448 RUS70261.1 KK853131 KDR10935.1 HACG01049256 CEK96121.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 RQTK01000357 RUS81065.1 RQTK01000058 RUS89376.1 KK852404 KDR24543.1 KK852941 KDR13536.1 GFAH01000222 JAV48167.1 RQTK01000752 RUS75316.1 RQTK01000360 RUS81011.1 NEVH01020333 PNF21801.1 KK853249 KDR09341.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
D5LB38
A0A2A4J795
A0A151IIH7
A0A146L0Z1
A0A2H1V1U0
A0A0K8T3U8
+ More
A0A2S2QJK3 A0A0A9XCF5 A0A0J7KRQ5 A0A0J7KZE8 A0A2A4JBK9 A0A2S2QAF2 A0A2S2PW26 A0A2W1BSK1 A0A0J7KK86 W4Y921 X1D4L4 A0A3S1AZH6 A0A3S0ZE77 J9LIJ7 A0A2S2P9Y7 A0A2S2P7C6 A0A3S1AY56 A0A067QXS5 A0A0B7BUS3 A0A0B7BSZ9 A0A0B7BT80 A0A0B7BUR9 A0A3S1A2L0 A0A3S1CD12 A0A067RNC1 A0A067QUH9 A0A1W7RAL1 A0A3S0ZU31 A0A3S1HJZ0 A0A2J7PZP5 A0A067QJX4
A0A2S2QJK3 A0A0A9XCF5 A0A0J7KRQ5 A0A0J7KZE8 A0A2A4JBK9 A0A2S2QAF2 A0A2S2PW26 A0A2W1BSK1 A0A0J7KK86 W4Y921 X1D4L4 A0A3S1AZH6 A0A3S0ZE77 J9LIJ7 A0A2S2P9Y7 A0A2S2P7C6 A0A3S1AY56 A0A067QXS5 A0A0B7BUS3 A0A0B7BSZ9 A0A0B7BT80 A0A0B7BUR9 A0A3S1A2L0 A0A3S1CD12 A0A067RNC1 A0A067QUH9 A0A1W7RAL1 A0A3S0ZU31 A0A3S1HJZ0 A0A2J7PZP5 A0A067QJX4
Ontologies
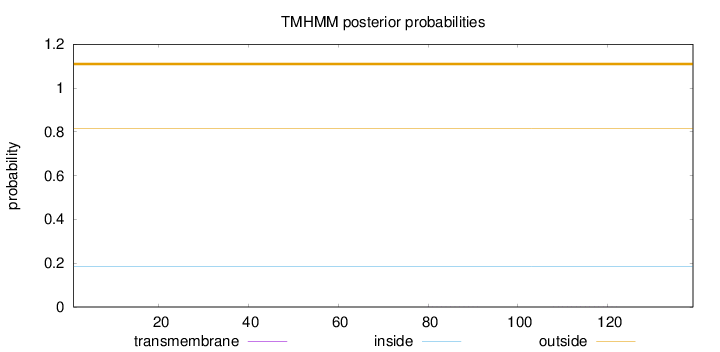
Topology
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0061
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.18445
outside
1 - 139
Population Genetic Test Statistics
Pi
315.567165
Theta
198.608789
Tajima's D
1.853572
CLR
0
CSRT
0.854057297135143
Interpretation
Uncertain
