Gene
KWMTBOMO05879
Pre Gene Modal
BGIBMGA002937
Annotation
PREDICTED:_nitric_oxide_synthase-like_protein_isoform_X1_[Bombyx_mori]
Full name
Nitric oxide synthase-like protein
+ More
Nitric oxide synthase
Nitric oxide synthase, salivary gland
Nitric oxide synthase
Nitric oxide synthase, salivary gland
Alternative Name
Inducible nitric oxide synthase-like protein
dNOS
dNOS
Location in the cell
Cytoplasmic Reliability : 1.854
Sequence
CDS
ATGTTCGAAGCACTCTGTAACCACATCAAGTACAGCACAAATAAAGGAAATATAAGATCAGCTATAACATTGTTCCCACAACGTACGGACGGCAAGCACGACTACAGAATATGGAACAGCCAGCTGATCAGCTACGCTGGCTACCGTCAGCCAGACGGTACGGTGCTTGGTGACCCCATGCACTGCGAATTCACTGATCTCTGCTTGAAACTTGGCTGGAAGCCGCCACGCACGGCCTGGGATATTCTTCCCCTAGTTCTGTCAGCTAACGGGAAAGACCCTGACTACTTCGAAATCCCCAGGGAGCTTGTTATGGAAATCCACATGACGCACCCCACGTTCGAATGGTTCAAAGAACTAGAATTACGTTGGTACGCTTTGCCCGCCGTCTCCAGTATGCGTTTTGATTGCGGTGGCATCGAATTCACGGCCAACGCTTTCAACGGTTGGTACATGAGCACTGAAATCGGCTGCAGGAACTTCTGTGACACCAACCGGCTTAATGTTTTGGAGAAAATAGCTCAAAACATGGGCCTAGATACCAGAACTCCGGTCAATCTTTGGAAGGATAAGGCCCTCGTGGAAGTGAACGTGGCTGTACTGCACAGCTTTCAACAGCACAACGCGACTATCGTGGATCATCATACAGCTTCTGAAAGCTTCATAAAGCATTTGGATAACGAGAACCGGCTTCGGTCTGGATGTCCAGCCGATTGGATATGGATCGTACCTCCCATGTCATCGTCCATAACCCCGGTGTTTCACCAAGAAATGGCGCTCTACTACTTGAAACCTTCATACGAATATCAGGAGCCTGCATGGAAGACGCATCAGTGGCAGAAATCTAAGCCGATCACAGGCAAAAGACCCATAAACAGAAAGTTCCATTTCAAGCAAATTGCCCGAGCTGTCAAATTCACTTCGAAATTGTTCGGTCGTGCCCTTTCGAAGAGGATTAAAGCGACTGTGCTGTACGCTACGGAGACTGGGAAATCGGAACAATACGCCAAAGAGCTTGGTGTTATATTTGGACACGCCTTTAATGCGCAGGTTCATTGTATGGCGGATTACGATATAACTTCAATTGAACACGAAGCTCTCCTTCTGGTTGTGACATCGACGTTTGGAAATGGTGACCCACCCGAGAATGGTGTCGCATTTGGAGAGCATCTATGCGAAATATTATACGCTGATCAAGTCGGTGAAGATTCTACTGGAAATCAAATGTTGACACCAAAATCGTTCATAAAAGCCAATAGTGACATTCAAAGGTATACAGCAGGAAATCCGAAAAAACTCAACAGACTCGAATCATTGAAGGGTAGTACAACAGACGCCACATCCATTGACAGCTTTGGTCCATTGAGTAATGTTAGATTTGCCGTGTTCGCTCTGGGTTCGAGCGCTTACCCGAATTTCTGTAACTTCGGAAAGTATGTAGACAAATTGTTGGTTGACCTCGGCGGGGAACGAATCCACGACCTCGCAACCGGTGACGAAATGTGCGGCCAAGATCAGGCCTTCCGGAAGTGGGCCTCTAGCGTCTTCAACGTTGCTTGTGAGACATTCTGCTTGGATGATGATGAGACTCTACAAGAGGCAAAGAGAGCTCTCGGTACTGTTGCATTAAGCGAGGAGACAGTTCAATTCGCCAGGGCTGGCTGTCGTCCCGCTACGTTACACGCAGCGCTACAAAAGTCTTTGAATAAACAATTCGTTTCTTGCACCGTCAAAGCTAATAAAGATTTAGGCGATGCTTCCGCTGAAAGATCCACCATCTTTATTGATCTGGAACCTAAGGAGGAAATAAAGTATAATCCAGGAGATCATGTCGGAATAATAGCTTGTAACCGTAAAGAGCTCGTTGAGAGTCTGCTCTCCCGCATTAAAGACGTTGATGATTACGACGAGCCGTTGCAGTTGCAGCTTCTGAAAGAAACTCATACGTCAAGCGGTTTAGTCAAATCTTGGGAGCCCCACGAGAAACTTCCGATAATGAGCGTGCGTGAATTGTTCACACGATTCCTTGATATAACAACTCCACCGACCACGATACTTCTTCAGTACTTGGCCACAACGTGCGAAGATGAAGAAGAAAAAAAACAATTGAATGTTTTGGCTACGGATCCCGGAGCTTACGAAGATTGGCGTCATTTCCACTTCCCCACCTTGCCCGAAGTCCTAGATCAGTTTCCATCAGCAAGACCCAATGCATCTTTACTTGCCGCCCTTTTATCCCCACTGCAGCCAAGATTTTACTCAATTTCTTCGTCACCATTAGCCCACGCAAAGAGATTGCATCTTACGGTTGCCGTAGTCACATATCGAACTCAAGATGGTGAGGGACCGGTCCATTATGGTGTGTGTTCCAACTACTTAATGGAACGTAAGCCTGGCGATGAGGTTTACCTCTTTATCAGAAGTGCTCCTAATTTCCATTTACCTCAAGATTTGTCAGTTCCTCTTATACTAATTGGTCCCGGTACTGGTATAGCTCCATTCCGTGGCTTTTGGCATCATCGGCGAGCTCTACAAAATTCTTGCTCGCGTACAACCACTGGACCGGTGTGGCTGTTCTTCGGCTGTCGAACGAAGACTATGGACTTATACCGAGAAGAGAAAGAGCAAGCTTTGAAAGAAGGCGTACTTTCTAAAGTATTTCTCGCTCTATCTAGAGAAAAGGAAGTTCCAAAAACGTACGTCCAAGAAGTTGCTGAAAACGTTGGTGCAGAAATACACGATTTACTAATTAATAAAGGTGCACACTTTTACGTTTGCGGTGATTGTAAAATGGCGGAGGATGTGCACCAGAAGCTCAAAGGTATCGTGAAGAAACACGGAAACATGACAGACGAACAAGTACAGAACTTTATGTTCATGCTAAAGGAAGAGAATCGTTACCACGAAGACATATTCGGCATCACACTGCGCACAGCTGAGGTCCACAGCGCTTCTCGCGAGTCGGCTCGTAGGAACCGCGTCGCTTCCCAACCATGA
Protein
MFEALCNHIKYSTNKGNIRSAITLFPQRTDGKHDYRIWNSQLISYAGYRQPDGTVLGDPMHCEFTDLCLKLGWKPPRTAWDILPLVLSANGKDPDYFEIPRELVMEIHMTHPTFEWFKELELRWYALPAVSSMRFDCGGIEFTANAFNGWYMSTEIGCRNFCDTNRLNVLEKIAQNMGLDTRTPVNLWKDKALVEVNVAVLHSFQQHNATIVDHHTASESFIKHLDNENRLRSGCPADWIWIVPPMSSSITPVFHQEMALYYLKPSYEYQEPAWKTHQWQKSKPITGKRPINRKFHFKQIARAVKFTSKLFGRALSKRIKATVLYATETGKSEQYAKELGVIFGHAFNAQVHCMADYDITSIEHEALLLVVTSTFGNGDPPENGVAFGEHLCEILYADQVGEDSTGNQMLTPKSFIKANSDIQRYTAGNPKKLNRLESLKGSTTDATSIDSFGPLSNVRFAVFALGSSAYPNFCNFGKYVDKLLVDLGGERIHDLATGDEMCGQDQAFRKWASSVFNVACETFCLDDDETLQEAKRALGTVALSEETVQFARAGCRPATLHAALQKSLNKQFVSCTVKANKDLGDASAERSTIFIDLEPKEEIKYNPGDHVGIIACNRKELVESLLSRIKDVDDYDEPLQLQLLKETHTSSGLVKSWEPHEKLPIMSVRELFTRFLDITTPPTTILLQYLATTCEDEEEKKQLNVLATDPGAYEDWRHFHFPTLPEVLDQFPSARPNASLLAALLSPLQPRFYSISSSPLAHAKRLHLTVAVVTYRTQDGEGPVHYGVCSNYLMERKPGDEVYLFIRSAPNFHLPQDLSVPLILIGPGTGIAPFRGFWHHRRALQNSCSRTTTGPVWLFFGCRTKTMDLYREEKEQALKEGVLSKVFLALSREKEVPKTYVQEVAENVGAEIHDLLINKGAHFYVCGDCKMAEDVHQKLKGIVKKHGNMTDEQVQNFMFMLKEENRYHEDIFGITLRTAEVHSASRESARRNRVASQP
Summary
Description
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body.
Produces nitric oxide (NO).
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Involved in the induction of immune gene expression.
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Nitric oxide limits plasmodium development in the midgut.
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Truncated isoforms (isoform 3-isoform 6) are able to form intracellular complexes with the full-length protein and serve as dominant negative inhibitors of the enzyme activity.
Produces nitric oxide (NO).
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Involved in the induction of immune gene expression.
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Nitric oxide limits plasmodium development in the midgut.
Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. Truncated isoforms (isoform 3-isoform 6) are able to form intracellular complexes with the full-length protein and serve as dominant negative inhibitors of the enzyme activity.
Catalytic Activity
H(+) + 2 L-arginine + 3 NADPH + 4 O2 = 4 H2O + 2 L-citrulline + 3 NADP(+) + 2 nitric oxide
Cofactor
heme
FAD
FMN
FAD
FMN
Similarity
Belongs to the NOS family.
Keywords
Alternative splicing
Calmodulin-binding
Complete proteome
FAD
Flavoprotein
FMN
Heme
Iron
Metal-binding
NADP
Oxidoreductase
Reference proteome
3D-structure
Feature
chain Nitric oxide synthase-like protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
B1B557
A0A3G2BAG7
A0A194QRD1
A0A2A4JPA3
A0A2W1BRS1
A0A2A4JQY7
+ More
A0A2H1VQU4 B1B557-2 A0A194Q9M0 A0A0L7L9D5 O77104 A0A194QQW5 A0A194Q846 Q8T8C0 A0A2J7R7T5 D6WZB5 A0A212FFL3 A0A2A4JQF6 A0A2W1BP40 A0A2A3EKD6 B7XH76 A0A2D1CSB0 Q5FAN1 A0A2H1WQB8 A0A310SCF2 A0A0L7R7J7 A0A0K0LIZ9 A0A0K0LJ45 A0A026VVR0 A0A0N0BC93 A0A154PDZ1 A5HVZ6 E9IZA1 A0A182UH37 A0A1S4GYE8 A0A195CZ09 Q7PNE2 A0A1Y9GKC1 A0A1Y9J085 A0A195AU06 A0A1S4GYH2 A5HVZ7 E2AAP2 A0A151WVV3 A0A151JCF9 A0A195FWG3 F4WUY7 A0A182PCN0 O61608 A0A182Y3I0 A0A1Y9IVD5 E2BIM4 A0A084W4T8 A0A182Q6X7 N6UIP3 Q16UY1 A0A218NGQ1 A0A336LEF7 A0A023FA12 E0VG92 A0A182V798 T1HIL3 A0A182L106 Q26240 H9TN28 W8BTS8 A0A034WS92 A0A2M4CJI3 A0A2M4BAZ3 D2YYH5 I2CZU9 A0A2M4CJ25 A0A2M4A368 D4N4H3 A0A2M4A372 A0A2M4A3A5 B4KKM7 A0A0M5IY53 D2J3S3 B4MU51 B4JPU3 A0A240SWJ7 G8XR43 A0A0U3BM43 A0A3B0JN27 A0A240SXC8 Q29KP6 B4LS74 Q27571 J9JN78 A0A1I8M1B8 A0A1W4W517 Q6E5V9 B4P121 A0A0R3P1B7 B3N527 G0M6G3 B3MJW2 A0A1I8Q4X2 B4HWW1
A0A2H1VQU4 B1B557-2 A0A194Q9M0 A0A0L7L9D5 O77104 A0A194QQW5 A0A194Q846 Q8T8C0 A0A2J7R7T5 D6WZB5 A0A212FFL3 A0A2A4JQF6 A0A2W1BP40 A0A2A3EKD6 B7XH76 A0A2D1CSB0 Q5FAN1 A0A2H1WQB8 A0A310SCF2 A0A0L7R7J7 A0A0K0LIZ9 A0A0K0LJ45 A0A026VVR0 A0A0N0BC93 A0A154PDZ1 A5HVZ6 E9IZA1 A0A182UH37 A0A1S4GYE8 A0A195CZ09 Q7PNE2 A0A1Y9GKC1 A0A1Y9J085 A0A195AU06 A0A1S4GYH2 A5HVZ7 E2AAP2 A0A151WVV3 A0A151JCF9 A0A195FWG3 F4WUY7 A0A182PCN0 O61608 A0A182Y3I0 A0A1Y9IVD5 E2BIM4 A0A084W4T8 A0A182Q6X7 N6UIP3 Q16UY1 A0A218NGQ1 A0A336LEF7 A0A023FA12 E0VG92 A0A182V798 T1HIL3 A0A182L106 Q26240 H9TN28 W8BTS8 A0A034WS92 A0A2M4CJI3 A0A2M4BAZ3 D2YYH5 I2CZU9 A0A2M4CJ25 A0A2M4A368 D4N4H3 A0A2M4A372 A0A2M4A3A5 B4KKM7 A0A0M5IY53 D2J3S3 B4MU51 B4JPU3 A0A240SWJ7 G8XR43 A0A0U3BM43 A0A3B0JN27 A0A240SXC8 Q29KP6 B4LS74 Q27571 J9JN78 A0A1I8M1B8 A0A1W4W517 Q6E5V9 B4P121 A0A0R3P1B7 B3N527 G0M6G3 B3MJW2 A0A1I8Q4X2 B4HWW1
EC Number
1.14.13.39
Pubmed
18505883
26354079
28756777
26227816
9736646
12000645
+ More
18362917 19820115 22118469 19437615 29035888 24508170 21282665 12364791 20798317 21719571 9576947 10333518 25244985 24438588 23537049 17510324 25474469 20566863 20966253 9022713 24495485 25348373 20026409 17994087 15632085 7568075 11526108 10731132 12537572 12804606 25315136 15235013 17550304
18362917 19820115 22118469 19437615 29035888 24508170 21282665 12364791 20798317 21719571 9576947 10333518 25244985 24438588 23537049 17510324 25474469 20566863 20966253 9022713 24495485 25348373 20026409 17994087 15632085 7568075 11526108 10731132 12537572 12804606 25315136 15235013 17550304
EMBL
AB360590
AB485776
BAG12563.1
BAH23564.1
MG770226
AYM26646.1
+ More
KQ461175 KPJ07899.1 NWSH01000856 PCG73831.1 KZ149998 PZC75366.1 PCG73832.1 ODYU01003891 SOQ43213.1 KQ459302 KPJ01710.1 JTDY01002100 KOB72118.1 AF062749 AAC61262.1 KPJ07898.1 KPJ01712.1 AB071182 AB485775 AB017521 BAB33296.1 BAB85836.1 BAH23563.1 NEVH01006727 PNF36893.1 KQ971372 EFA10400.2 AGBW02008807 OWR52517.1 PCG73834.1 PZC75365.1 KZ288232 PBC31491.1 AB477987 BAH14964.1 MF162295 ATN45429.1 AB204558 BAD89803.1 ODYU01010272 SOQ55265.1 KQ761346 OAD57843.1 KQ414639 KOC66824.1 KJ425527 AJA40862.1 KJ425525 AJA40859.1 KK107770 EZA47847.1 KQ435922 KOX68681.1 KQ434874 KZC09634.1 AB304919 BAF63160.1 GL767121 EFZ14055.1 KQ977141 KYN05369.1 AAAB01008964 EAA12335.2 APCN01000801 APCN01000802 KQ976738 KYM75666.1 AB304920 BAF63161.1 GL438137 EFN69492.1 KQ982691 KYQ52059.1 KQ979038 KYN23479.1 KQ981200 KYN45005.1 GL888378 EGI62001.1 AF130134 AF130124 AF130125 AF130126 AF130127 AF130128 AF130129 AF130130 AF130131 AF130132 AF130133 GL448516 EFN84471.1 ATLV01020382 ATLV01020383 KE525299 KFB45232.1 AXCN02000784 APGK01018684 APGK01018685 APGK01018686 APGK01018687 APGK01018688 APGK01018689 APGK01018690 APGK01018691 KB740082 ENN81585.1 CH477609 EAT38354.1 KY465473 ASG81432.1 UFQS01002944 UFQT01002944 SSX14935.1 SSX34322.1 GBBI01000949 JAC17763.1 DS235133 EEB12398.1 ACPB03007893 U59389 JF502067 AFG22596.1 GAMC01014036 JAB92519.1 GAKP01002324 GAKP01002323 JAC56628.1 GGFL01001306 MBW65484.1 GGFJ01000837 MBW49978.1 AB485762 BAI67609.1 JQ793785 AFJ74715.1 GGFL01001077 MBW65255.1 GGFK01001851 MBW35172.1 GQ429217 ADD63793.1 GGFK01001849 MBW35170.1 GGFK01001946 MBW35267.1 CH933807 EDW12691.1 CP012523 ALC38686.1 GQ865598 ACZ60615.1 CH963852 EDW75640.1 CH916372 EDV98923.1 EU863412 ACJ54486.1 KU306112 ALT10385.1 OUUW01000006 SPP81742.1 JXJN01001049 JXJN01001050 JXJN01001051 JXJN01001052 JXJN01001053 JXJN01001054 JXJN01001055 JXJN01001056 JXJN01001057 JXJN01001058 CH379061 EAL33128.1 CH940649 EDW64760.2 U25117 AF215700 AF215691 AF215692 AF215693 AF215694 AF215695 AF215696 AF215697 AF215698 AF215699 AE014134 BT010016 ABLF02036032 ABLF02036036 ABLF02036037 AY552549 AAT46681.1 CM000157 EDW87996.1 KRT04758.1 CH954177 EDV58972.1 FR875156 CCC18661.1 CH902620 EDV32417.1 CH480818 EDW52506.1
KQ461175 KPJ07899.1 NWSH01000856 PCG73831.1 KZ149998 PZC75366.1 PCG73832.1 ODYU01003891 SOQ43213.1 KQ459302 KPJ01710.1 JTDY01002100 KOB72118.1 AF062749 AAC61262.1 KPJ07898.1 KPJ01712.1 AB071182 AB485775 AB017521 BAB33296.1 BAB85836.1 BAH23563.1 NEVH01006727 PNF36893.1 KQ971372 EFA10400.2 AGBW02008807 OWR52517.1 PCG73834.1 PZC75365.1 KZ288232 PBC31491.1 AB477987 BAH14964.1 MF162295 ATN45429.1 AB204558 BAD89803.1 ODYU01010272 SOQ55265.1 KQ761346 OAD57843.1 KQ414639 KOC66824.1 KJ425527 AJA40862.1 KJ425525 AJA40859.1 KK107770 EZA47847.1 KQ435922 KOX68681.1 KQ434874 KZC09634.1 AB304919 BAF63160.1 GL767121 EFZ14055.1 KQ977141 KYN05369.1 AAAB01008964 EAA12335.2 APCN01000801 APCN01000802 KQ976738 KYM75666.1 AB304920 BAF63161.1 GL438137 EFN69492.1 KQ982691 KYQ52059.1 KQ979038 KYN23479.1 KQ981200 KYN45005.1 GL888378 EGI62001.1 AF130134 AF130124 AF130125 AF130126 AF130127 AF130128 AF130129 AF130130 AF130131 AF130132 AF130133 GL448516 EFN84471.1 ATLV01020382 ATLV01020383 KE525299 KFB45232.1 AXCN02000784 APGK01018684 APGK01018685 APGK01018686 APGK01018687 APGK01018688 APGK01018689 APGK01018690 APGK01018691 KB740082 ENN81585.1 CH477609 EAT38354.1 KY465473 ASG81432.1 UFQS01002944 UFQT01002944 SSX14935.1 SSX34322.1 GBBI01000949 JAC17763.1 DS235133 EEB12398.1 ACPB03007893 U59389 JF502067 AFG22596.1 GAMC01014036 JAB92519.1 GAKP01002324 GAKP01002323 JAC56628.1 GGFL01001306 MBW65484.1 GGFJ01000837 MBW49978.1 AB485762 BAI67609.1 JQ793785 AFJ74715.1 GGFL01001077 MBW65255.1 GGFK01001851 MBW35172.1 GQ429217 ADD63793.1 GGFK01001849 MBW35170.1 GGFK01001946 MBW35267.1 CH933807 EDW12691.1 CP012523 ALC38686.1 GQ865598 ACZ60615.1 CH963852 EDW75640.1 CH916372 EDV98923.1 EU863412 ACJ54486.1 KU306112 ALT10385.1 OUUW01000006 SPP81742.1 JXJN01001049 JXJN01001050 JXJN01001051 JXJN01001052 JXJN01001053 JXJN01001054 JXJN01001055 JXJN01001056 JXJN01001057 JXJN01001058 CH379061 EAL33128.1 CH940649 EDW64760.2 U25117 AF215700 AF215691 AF215692 AF215693 AF215694 AF215695 AF215696 AF215697 AF215698 AF215699 AE014134 BT010016 ABLF02036032 ABLF02036036 ABLF02036037 AY552549 AAT46681.1 CM000157 EDW87996.1 KRT04758.1 CH954177 EDV58972.1 FR875156 CCC18661.1 CH902620 EDV32417.1 CH480818 EDW52506.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000037510
UP000235965
+ More
UP000007266 UP000007151 UP000242457 UP000005203 UP000053825 UP000053097 UP000053105 UP000076502 UP000075902 UP000078542 UP000007062 UP000075840 UP000076407 UP000078540 UP000000311 UP000075809 UP000078492 UP000078541 UP000007755 UP000075885 UP000076408 UP000075920 UP000008237 UP000030765 UP000075886 UP000019118 UP000008820 UP000009046 UP000075903 UP000015103 UP000075882 UP000009192 UP000092553 UP000007798 UP000001070 UP000092443 UP000268350 UP000092460 UP000001819 UP000008792 UP000000803 UP000007819 UP000095301 UP000192221 UP000002282 UP000008711 UP000007801 UP000095300 UP000001292
UP000007266 UP000007151 UP000242457 UP000005203 UP000053825 UP000053097 UP000053105 UP000076502 UP000075902 UP000078542 UP000007062 UP000075840 UP000076407 UP000078540 UP000000311 UP000075809 UP000078492 UP000078541 UP000007755 UP000075885 UP000076408 UP000075920 UP000008237 UP000030765 UP000075886 UP000019118 UP000008820 UP000009046 UP000075903 UP000015103 UP000075882 UP000009192 UP000092553 UP000007798 UP000001070 UP000092443 UP000268350 UP000092460 UP000001819 UP000008792 UP000000803 UP000007819 UP000095301 UP000192221 UP000002282 UP000008711 UP000007801 UP000095300 UP000001292
Interpro
IPR001709
Flavoprot_Pyr_Nucl_cyt_Rdtase
+ More
IPR039261 FNR_nucleotide-bd
IPR003097 CysJ-like_FAD-binding
IPR036119 NOS_N_sf
IPR029039 Flavoprotein-like_sf
IPR012144 NOS_euk
IPR001094 Flavdoxin-like
IPR008254 Flavodoxin/NO_synth
IPR001433 OxRdtase_FAD/NAD-bd
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR017927 FAD-bd_FR_type
IPR004030 NOS_N
IPR017938 Riboflavin_synthase-like_b-brl
IPR039261 FNR_nucleotide-bd
IPR003097 CysJ-like_FAD-binding
IPR036119 NOS_N_sf
IPR029039 Flavoprotein-like_sf
IPR012144 NOS_euk
IPR001094 Flavdoxin-like
IPR008254 Flavodoxin/NO_synth
IPR001433 OxRdtase_FAD/NAD-bd
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR017927 FAD-bd_FR_type
IPR004030 NOS_N
IPR017938 Riboflavin_synthase-like_b-brl
Gene 3D
ProteinModelPortal
B1B557
A0A3G2BAG7
A0A194QRD1
A0A2A4JPA3
A0A2W1BRS1
A0A2A4JQY7
+ More
A0A2H1VQU4 B1B557-2 A0A194Q9M0 A0A0L7L9D5 O77104 A0A194QQW5 A0A194Q846 Q8T8C0 A0A2J7R7T5 D6WZB5 A0A212FFL3 A0A2A4JQF6 A0A2W1BP40 A0A2A3EKD6 B7XH76 A0A2D1CSB0 Q5FAN1 A0A2H1WQB8 A0A310SCF2 A0A0L7R7J7 A0A0K0LIZ9 A0A0K0LJ45 A0A026VVR0 A0A0N0BC93 A0A154PDZ1 A5HVZ6 E9IZA1 A0A182UH37 A0A1S4GYE8 A0A195CZ09 Q7PNE2 A0A1Y9GKC1 A0A1Y9J085 A0A195AU06 A0A1S4GYH2 A5HVZ7 E2AAP2 A0A151WVV3 A0A151JCF9 A0A195FWG3 F4WUY7 A0A182PCN0 O61608 A0A182Y3I0 A0A1Y9IVD5 E2BIM4 A0A084W4T8 A0A182Q6X7 N6UIP3 Q16UY1 A0A218NGQ1 A0A336LEF7 A0A023FA12 E0VG92 A0A182V798 T1HIL3 A0A182L106 Q26240 H9TN28 W8BTS8 A0A034WS92 A0A2M4CJI3 A0A2M4BAZ3 D2YYH5 I2CZU9 A0A2M4CJ25 A0A2M4A368 D4N4H3 A0A2M4A372 A0A2M4A3A5 B4KKM7 A0A0M5IY53 D2J3S3 B4MU51 B4JPU3 A0A240SWJ7 G8XR43 A0A0U3BM43 A0A3B0JN27 A0A240SXC8 Q29KP6 B4LS74 Q27571 J9JN78 A0A1I8M1B8 A0A1W4W517 Q6E5V9 B4P121 A0A0R3P1B7 B3N527 G0M6G3 B3MJW2 A0A1I8Q4X2 B4HWW1
A0A2H1VQU4 B1B557-2 A0A194Q9M0 A0A0L7L9D5 O77104 A0A194QQW5 A0A194Q846 Q8T8C0 A0A2J7R7T5 D6WZB5 A0A212FFL3 A0A2A4JQF6 A0A2W1BP40 A0A2A3EKD6 B7XH76 A0A2D1CSB0 Q5FAN1 A0A2H1WQB8 A0A310SCF2 A0A0L7R7J7 A0A0K0LIZ9 A0A0K0LJ45 A0A026VVR0 A0A0N0BC93 A0A154PDZ1 A5HVZ6 E9IZA1 A0A182UH37 A0A1S4GYE8 A0A195CZ09 Q7PNE2 A0A1Y9GKC1 A0A1Y9J085 A0A195AU06 A0A1S4GYH2 A5HVZ7 E2AAP2 A0A151WVV3 A0A151JCF9 A0A195FWG3 F4WUY7 A0A182PCN0 O61608 A0A182Y3I0 A0A1Y9IVD5 E2BIM4 A0A084W4T8 A0A182Q6X7 N6UIP3 Q16UY1 A0A218NGQ1 A0A336LEF7 A0A023FA12 E0VG92 A0A182V798 T1HIL3 A0A182L106 Q26240 H9TN28 W8BTS8 A0A034WS92 A0A2M4CJI3 A0A2M4BAZ3 D2YYH5 I2CZU9 A0A2M4CJ25 A0A2M4A368 D4N4H3 A0A2M4A372 A0A2M4A3A5 B4KKM7 A0A0M5IY53 D2J3S3 B4MU51 B4JPU3 A0A240SWJ7 G8XR43 A0A0U3BM43 A0A3B0JN27 A0A240SXC8 Q29KP6 B4LS74 Q27571 J9JN78 A0A1I8M1B8 A0A1W4W517 Q6E5V9 B4P121 A0A0R3P1B7 B3N527 G0M6G3 B3MJW2 A0A1I8Q4X2 B4HWW1
PDB
1TLL
E-value=1.12615e-147,
Score=1345
Ontologies
GO
GO:0050660
GO:0006809
GO:0050661
GO:0004517
GO:0020037
GO:0005516
GO:0010181
GO:0046872
GO:0016491
GO:0005829
GO:0032496
GO:0012506
GO:0031284
GO:0005634
GO:0009725
GO:0005886
GO:0006527
GO:0007263
GO:0003958
GO:0002027
GO:0007399
GO:0007444
GO:0046620
GO:0008156
GO:0005777
GO:0007416
GO:0008285
GO:0005506
GO:0016597
GO:0055114
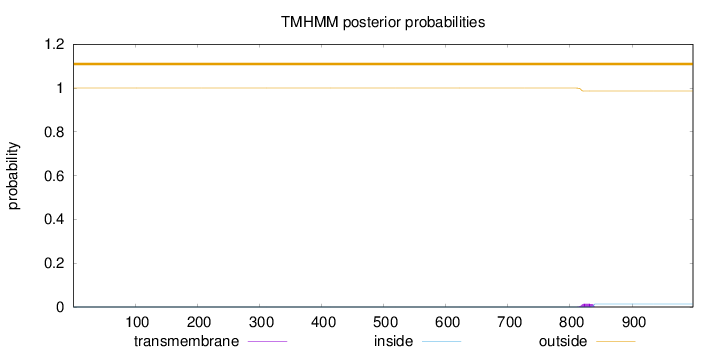
Topology
Length:
998
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29119
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 998
Population Genetic Test Statistics
Pi
213.531004
Theta
170.980429
Tajima's D
0.714031
CLR
0.065279
CSRT
0.574421278936053
Interpretation
Uncertain
