Gene
KWMTBOMO05874 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002939
Annotation
PREDICTED:_15-hydroxyprostaglandin_dehydrogenase_[NAD(+)]-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.496
Sequence
CDS
ATGTCGCACGAATGGAAAGATAAAGTAGTTTTTATAACCGGTGCTGCAACGGGCATCGGTGAATCTGTGGTCAGGATATTATTAGACGAAGGAGTTAAGCATATAGCGATTTTGGACATCGCGGAAGAAGCCGGACAATCGTTACAAGCTGAACTGAATTCGAAATATGGCAACAAAACGAAGTTCTACAGGTCTGACGTCACTGATGAAGAGCACTTCCTGGGAATCCTTAACGCTGTAGTTCAGGAGCAAGGCGGCATTGATGTAGTGATTAATAATGCCGCCCTCATGAACGACAGTATTGGCATTTACAAAAAAGCCATTGAGGTCAACGTGACCGCCTTAATAACAAGTACATTAAAAGCCATTGAGATAATGGGGAAGCACAGCGGTGGTAAAGGAGGTACAGTTATCAACGTTTCTTCGGTTGCCGCTTTGACGCAATCTCACGTTATGCCAATATACTTCGCAACTAAAAGCGCTGTGCTGCAGTTCAGTAATTGTATCGGTAGACCGGAACACTTTTCGAAAACCGGGGTCCGAGTACTGACAGTGTGCTTTGGTGCAACAGACACCGCGTTGTTGACCGACGAGAAACTAGGCACTTTTGATAAGTACTTAGAATCTTCATGCTTGGACCAACTAAAGAAAGGCTTCATATTCCAACGCAAAGAATCCGCAGCTCAAGGCCTCGTTGATTCTTACAAGAAAGGTGAGAGTGGCAGTACATGGCTCGTTGTAAATAACTTGCCGGTTCAAGAGATCAGCGGGTCGGTCGCCGAAGCCTATCAGATTCTTTCTCAGAATGTTTATAGACTTTAA
Protein
MSHEWKDKVVFITGAATGIGESVVRILLDEGVKHIAILDIAEEAGQSLQAELNSKYGNKTKFYRSDVTDEEHFLGILNAVVQEQGGIDVVINNAALMNDSIGIYKKAIEVNVTALITSTLKAIEIMGKHSGGKGGTVINVSSVAALTQSHVMPIYFATKSAVLQFSNCIGRPEHFSKTGVRVLTVCFGATDTALLTDEKLGTFDKYLESSCLDQLKKGFIFQRKESAAQGLVDSYKKGESGSTWLVVNNLPVQEISGSVAEAYQILSQNVYRL
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J0A3
H9J0A5
H9J0A4
A0A3G1T1B2
A0A0H4TG49
A0A212F0I1
+ More
A0A212F0G8 A0A194QE27 A0A194QRC6 A0A212F0G1 A0A076FT24 A0A0F6Q4J7 A0A1L8D6X2 A0A194Q8J5 A0A3S2PLF3 A0A194QQX1 A0A0L7KWX5 H9J0A6 H9J0A8 A0A3S2LVR9 A0A2W1BM47 A0A0L7KNY8 A0A212F0F9 A0A0L7KXH6 A0A3S2NMK6 A0A0L7KNH1 A0A0N0PD11 U5KCB3 A0A2H1VGZ5 A0A2H1VH14 A0A194QFX2 A0A2A4JLC0 A0A2W1BJY5 A0A2A4K9E1 A0A2H1VGZ4 H9J0A9 Q1G151 A0A194Q851 A0A194QQW0 A0A2H1WQ20 A0A0N1IPP9 A0A194QA61 A0A2H1WWP5 A0A194QA14 A0A0H4TY65 A0A2H1W7D8 A0A2A4JMW2 A0A2A4KAR2 A0A0F6Q2Y5 A0A2A4JMY9 A0A0N1PH34 A0A0N0PDG0 A0A224XI56 Q6RUR3 A0A026WRA5 A0A1Y1MAW1 A0A069DY58 A0A0J7L0X1 A0A0N0PD56 A0A067QWG1 A0A195E5P2 A0A023F7A3 F4W475 A0A151WLU8 E2A712 A0A026WSD7 H9JQQ4 A0A310S8E6 A0A195B3G3 A0A151X250 I6QWT6 A0A0L7QT43 A0A224XI78 A0A1W4X358 K7IV86 A0A195EUB2 A0A1W4XDZ2 A0A0L7L646 A0A194R4F0 A0A151XEI7 A0A195CJP6 G8JKE8 A0A0K8S9E4 A0A2J7QGZ8 A0A088A501 A0A0A9WS12 A0A232EGH1 A0A195ET92 A0A0J7L1B2 A0A212F936 E2B6N1 E0VAP2 E9I9W8 A0A154PC81 A0A158NDP0
A0A212F0G8 A0A194QE27 A0A194QRC6 A0A212F0G1 A0A076FT24 A0A0F6Q4J7 A0A1L8D6X2 A0A194Q8J5 A0A3S2PLF3 A0A194QQX1 A0A0L7KWX5 H9J0A6 H9J0A8 A0A3S2LVR9 A0A2W1BM47 A0A0L7KNY8 A0A212F0F9 A0A0L7KXH6 A0A3S2NMK6 A0A0L7KNH1 A0A0N0PD11 U5KCB3 A0A2H1VGZ5 A0A2H1VH14 A0A194QFX2 A0A2A4JLC0 A0A2W1BJY5 A0A2A4K9E1 A0A2H1VGZ4 H9J0A9 Q1G151 A0A194Q851 A0A194QQW0 A0A2H1WQ20 A0A0N1IPP9 A0A194QA61 A0A2H1WWP5 A0A194QA14 A0A0H4TY65 A0A2H1W7D8 A0A2A4JMW2 A0A2A4KAR2 A0A0F6Q2Y5 A0A2A4JMY9 A0A0N1PH34 A0A0N0PDG0 A0A224XI56 Q6RUR3 A0A026WRA5 A0A1Y1MAW1 A0A069DY58 A0A0J7L0X1 A0A0N0PD56 A0A067QWG1 A0A195E5P2 A0A023F7A3 F4W475 A0A151WLU8 E2A712 A0A026WSD7 H9JQQ4 A0A310S8E6 A0A195B3G3 A0A151X250 I6QWT6 A0A0L7QT43 A0A224XI78 A0A1W4X358 K7IV86 A0A195EUB2 A0A1W4XDZ2 A0A0L7L646 A0A194R4F0 A0A151XEI7 A0A195CJP6 G8JKE8 A0A0K8S9E4 A0A2J7QGZ8 A0A088A501 A0A0A9WS12 A0A232EGH1 A0A195ET92 A0A0J7L1B2 A0A212F936 E2B6N1 E0VAP2 E9I9W8 A0A154PC81 A0A158NDP0
Pubmed
EMBL
BABH01019200
BABH01019208
BABH01019201
BABH01019202
MG846919
AXY94771.1
+ More
KP899548 AKQ06147.1 AGBW02011082 OWR47240.1 OWR47239.1 KQ459302 KPJ01716.1 KQ461175 KPJ07894.1 OWR47236.1 KF960797 AII21999.1 KP237895 AKD01748.1 GEYN01000012 JAV02117.1 KPJ01719.1 RSAL01000003 RVE54802.1 KPJ07892.1 JTDY01004799 KOB67737.1 BABH01019209 BABH01019210 RVE54801.1 KZ149998 PZC75361.1 JTDY01007982 KOB64820.1 OWR47238.1 KOB67739.1 RVE54803.1 JTDY01007968 KOB64832.1 KQ460366 KPJ15584.1 KC007341 AGQ45606.1 ODYU01002522 SOQ40120.1 SOQ40118.1 KQ459249 KPJ02346.1 NWSH01001071 PCG72771.1 PZC75359.1 NWSH01000014 PCG80831.1 SOQ40119.1 BABH01019219 DQ512730 ABF59817.1 KPJ01717.1 KPJ07893.1 ODYU01010208 SOQ55170.1 KPJ15603.1 KPJ02362.1 ODYU01011579 SOQ57407.1 KPJ02363.1 KP899552 AKQ06151.1 ODYU01006680 SOQ48742.1 PCG72773.1 PCG80830.1 KP237897 AKD01750.1 PCG72772.1 KPJ15583.1 KPJ15602.1 GFTR01004259 JAW12167.1 AY491503 AAR84629.1 KK107119 QOIP01000009 EZA58570.1 RLU18591.1 GEZM01036109 GEZM01036108 JAV82969.1 GBGD01002580 JAC86309.1 LBMM01001364 KMQ96442.1 KPJ15601.1 KK852879 KDR14474.1 KQ979609 KYN20169.1 GBBI01001633 JAC17079.1 GL887491 EGI71071.1 KQ982944 KYQ48859.1 GL437252 EFN70744.1 EZA58571.1 RLU18590.1 BABH01012284 BABH01012285 KQ771134 OAD52541.1 KQ976625 KYM79033.1 KQ982585 KYQ54300.1 JQ398675 AFM38190.1 KQ414755 KOC61724.1 GFTR01004241 JAW12185.1 AAZX01008191 KQ981979 KYN31469.1 JTDY01002703 KOB70885.1 KQ460779 KPJ12399.1 KQ982254 KYQ58773.1 KQ977642 KYN00938.1 JP593077 AER92462.1 GBRD01015928 JAG49898.1 NEVH01014359 PNF27833.1 GBHO01034331 GDHC01017826 JAG09273.1 JAQ00803.1 NNAY01004765 OXU17447.1 KYN31470.1 KMQ96441.1 AGBW02009651 OWR50237.1 GL446000 EFN88615.1 DS235012 EEB10448.1 GL761846 EFZ22664.1 KQ434870 KZC09506.1 ADTU01012731
KP899548 AKQ06147.1 AGBW02011082 OWR47240.1 OWR47239.1 KQ459302 KPJ01716.1 KQ461175 KPJ07894.1 OWR47236.1 KF960797 AII21999.1 KP237895 AKD01748.1 GEYN01000012 JAV02117.1 KPJ01719.1 RSAL01000003 RVE54802.1 KPJ07892.1 JTDY01004799 KOB67737.1 BABH01019209 BABH01019210 RVE54801.1 KZ149998 PZC75361.1 JTDY01007982 KOB64820.1 OWR47238.1 KOB67739.1 RVE54803.1 JTDY01007968 KOB64832.1 KQ460366 KPJ15584.1 KC007341 AGQ45606.1 ODYU01002522 SOQ40120.1 SOQ40118.1 KQ459249 KPJ02346.1 NWSH01001071 PCG72771.1 PZC75359.1 NWSH01000014 PCG80831.1 SOQ40119.1 BABH01019219 DQ512730 ABF59817.1 KPJ01717.1 KPJ07893.1 ODYU01010208 SOQ55170.1 KPJ15603.1 KPJ02362.1 ODYU01011579 SOQ57407.1 KPJ02363.1 KP899552 AKQ06151.1 ODYU01006680 SOQ48742.1 PCG72773.1 PCG80830.1 KP237897 AKD01750.1 PCG72772.1 KPJ15583.1 KPJ15602.1 GFTR01004259 JAW12167.1 AY491503 AAR84629.1 KK107119 QOIP01000009 EZA58570.1 RLU18591.1 GEZM01036109 GEZM01036108 JAV82969.1 GBGD01002580 JAC86309.1 LBMM01001364 KMQ96442.1 KPJ15601.1 KK852879 KDR14474.1 KQ979609 KYN20169.1 GBBI01001633 JAC17079.1 GL887491 EGI71071.1 KQ982944 KYQ48859.1 GL437252 EFN70744.1 EZA58571.1 RLU18590.1 BABH01012284 BABH01012285 KQ771134 OAD52541.1 KQ976625 KYM79033.1 KQ982585 KYQ54300.1 JQ398675 AFM38190.1 KQ414755 KOC61724.1 GFTR01004241 JAW12185.1 AAZX01008191 KQ981979 KYN31469.1 JTDY01002703 KOB70885.1 KQ460779 KPJ12399.1 KQ982254 KYQ58773.1 KQ977642 KYN00938.1 JP593077 AER92462.1 GBRD01015928 JAG49898.1 NEVH01014359 PNF27833.1 GBHO01034331 GDHC01017826 JAG09273.1 JAQ00803.1 NNAY01004765 OXU17447.1 KYN31470.1 KMQ96441.1 AGBW02009651 OWR50237.1 GL446000 EFN88615.1 DS235012 EEB10448.1 GL761846 EFZ22664.1 KQ434870 KZC09506.1 ADTU01012731
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000283053
UP000037510
+ More
UP000218220 UP000053097 UP000279307 UP000036403 UP000027135 UP000078492 UP000007755 UP000075809 UP000000311 UP000078540 UP000053825 UP000192223 UP000002358 UP000078541 UP000078542 UP000235965 UP000005203 UP000215335 UP000008237 UP000009046 UP000076502 UP000005205
UP000218220 UP000053097 UP000279307 UP000036403 UP000027135 UP000078492 UP000007755 UP000075809 UP000000311 UP000078540 UP000053825 UP000192223 UP000002358 UP000078541 UP000078542 UP000235965 UP000005203 UP000215335 UP000008237 UP000009046 UP000076502 UP000005205
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J0A3
H9J0A5
H9J0A4
A0A3G1T1B2
A0A0H4TG49
A0A212F0I1
+ More
A0A212F0G8 A0A194QE27 A0A194QRC6 A0A212F0G1 A0A076FT24 A0A0F6Q4J7 A0A1L8D6X2 A0A194Q8J5 A0A3S2PLF3 A0A194QQX1 A0A0L7KWX5 H9J0A6 H9J0A8 A0A3S2LVR9 A0A2W1BM47 A0A0L7KNY8 A0A212F0F9 A0A0L7KXH6 A0A3S2NMK6 A0A0L7KNH1 A0A0N0PD11 U5KCB3 A0A2H1VGZ5 A0A2H1VH14 A0A194QFX2 A0A2A4JLC0 A0A2W1BJY5 A0A2A4K9E1 A0A2H1VGZ4 H9J0A9 Q1G151 A0A194Q851 A0A194QQW0 A0A2H1WQ20 A0A0N1IPP9 A0A194QA61 A0A2H1WWP5 A0A194QA14 A0A0H4TY65 A0A2H1W7D8 A0A2A4JMW2 A0A2A4KAR2 A0A0F6Q2Y5 A0A2A4JMY9 A0A0N1PH34 A0A0N0PDG0 A0A224XI56 Q6RUR3 A0A026WRA5 A0A1Y1MAW1 A0A069DY58 A0A0J7L0X1 A0A0N0PD56 A0A067QWG1 A0A195E5P2 A0A023F7A3 F4W475 A0A151WLU8 E2A712 A0A026WSD7 H9JQQ4 A0A310S8E6 A0A195B3G3 A0A151X250 I6QWT6 A0A0L7QT43 A0A224XI78 A0A1W4X358 K7IV86 A0A195EUB2 A0A1W4XDZ2 A0A0L7L646 A0A194R4F0 A0A151XEI7 A0A195CJP6 G8JKE8 A0A0K8S9E4 A0A2J7QGZ8 A0A088A501 A0A0A9WS12 A0A232EGH1 A0A195ET92 A0A0J7L1B2 A0A212F936 E2B6N1 E0VAP2 E9I9W8 A0A154PC81 A0A158NDP0
A0A212F0G8 A0A194QE27 A0A194QRC6 A0A212F0G1 A0A076FT24 A0A0F6Q4J7 A0A1L8D6X2 A0A194Q8J5 A0A3S2PLF3 A0A194QQX1 A0A0L7KWX5 H9J0A6 H9J0A8 A0A3S2LVR9 A0A2W1BM47 A0A0L7KNY8 A0A212F0F9 A0A0L7KXH6 A0A3S2NMK6 A0A0L7KNH1 A0A0N0PD11 U5KCB3 A0A2H1VGZ5 A0A2H1VH14 A0A194QFX2 A0A2A4JLC0 A0A2W1BJY5 A0A2A4K9E1 A0A2H1VGZ4 H9J0A9 Q1G151 A0A194Q851 A0A194QQW0 A0A2H1WQ20 A0A0N1IPP9 A0A194QA61 A0A2H1WWP5 A0A194QA14 A0A0H4TY65 A0A2H1W7D8 A0A2A4JMW2 A0A2A4KAR2 A0A0F6Q2Y5 A0A2A4JMY9 A0A0N1PH34 A0A0N0PDG0 A0A224XI56 Q6RUR3 A0A026WRA5 A0A1Y1MAW1 A0A069DY58 A0A0J7L0X1 A0A0N0PD56 A0A067QWG1 A0A195E5P2 A0A023F7A3 F4W475 A0A151WLU8 E2A712 A0A026WSD7 H9JQQ4 A0A310S8E6 A0A195B3G3 A0A151X250 I6QWT6 A0A0L7QT43 A0A224XI78 A0A1W4X358 K7IV86 A0A195EUB2 A0A1W4XDZ2 A0A0L7L646 A0A194R4F0 A0A151XEI7 A0A195CJP6 G8JKE8 A0A0K8S9E4 A0A2J7QGZ8 A0A088A501 A0A0A9WS12 A0A232EGH1 A0A195ET92 A0A0J7L1B2 A0A212F936 E2B6N1 E0VAP2 E9I9W8 A0A154PC81 A0A158NDP0
PDB
2GDZ
E-value=4.84231e-26,
Score=291
Ontologies
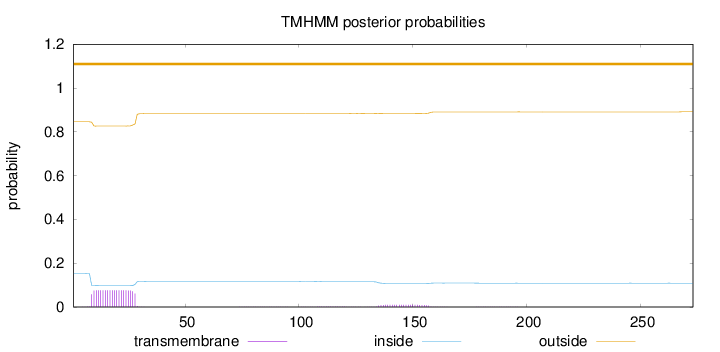
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.75598
Exp number, first 60 AAs:
1.49556
Total prob of N-in:
0.15443
outside
1 - 273
Population Genetic Test Statistics
Pi
182.331363
Theta
170.838367
Tajima's D
0.533114
CLR
0.444464
CSRT
0.518574071296435
Interpretation
Uncertain
