Gene
KWMTBOMO05865
Pre Gene Modal
BGIBMGA002945
Annotation
putative_alcohol_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.329
Sequence
CDS
ATGGCATCGGATTTCGTGAAGCGTTTTGTGGATGTCGATGATAAAGTGTTCTTGGTGACCGGTGGTGCGGCCGGCGTAGGCGCTGGCTTGGTCAAGGCGTTGCTGTTCGAAAACGCACGGCACGTCGCGTTTCTAGATGTCGCTGACCGAGAAGGGGCCGCCCTGGAGGCCCAACTTATAATCAAGTTCGGTGCCTTGAGAGTGAAGTTCATAAAGTGCGACGTAGGAGACGAACGTCAACTCGCTTCTGCATACAAACAAGTGTTGGACAAATACAAAAGGCTTGACGGCGTCATAAACAGCGCTGCAGTACTTAGCGTCGATGATAACTCTTTCAACAGAATGATTGATATCAATTTTACGGGCACAGTGAATAGTACTCTGAAAGCGCTCGATATTATGGGGGCAGATAAAGGCGGTTCTGGCGGTGTTATAGTGAATATATCGTCCCTACTTGCTCTTAATCTCACCAGCCATTTACCTGTTTACGCCGCTACTAAAGCAGCGGTATTACAGTTCAGCATTAGAATGGGCACGCAGGAACAATTTACCCGGACTAAAGTACGGGTATTGTCCGTATGCCTCGGGCCCACTGACACTGCAATCCTTTACAAGAACAACTTAACCAAGTTTGATACAGAGTACGCTCCGTGTTTGTCTTCGCGTGCCCCCGTGAGACAAAGAGTCGAGTCAGCTGTTAAGGGTATTTTGGAGGTGATCAACAAAGCGAGCACCGGAGATACGTGGATCGTGGAAAGCGACAAACCGGCTTACGATTTTACTCAAAATATATCCGAAGCCTTCCAAATACTCTCCAAGACATAG
Protein
MASDFVKRFVDVDDKVFLVTGGAAGVGAGLVKALLFENARHVAFLDVADREGAALEAQLIIKFGALRVKFIKCDVGDERQLASAYKQVLDKYKRLDGVINSAAVLSVDDNSFNRMIDINFTGTVNSTLKALDIMGADKGGSGGVIVNISSLLALNLTSHLPVYAATKAAVLQFSIRMGTQEQFTRTKVRVLSVCLGPTDTAILYKNNLTKFDTEYAPCLSSRAPVRQRVESAVKGILEVINKASTGDTWIVESDKPAYDFTQNISEAFQILSKT
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J0A9
Q1G151
A0A2W1BJY5
A0A2A4K9E1
A0A2A4KAR2
A0A0H4TY65
+ More
A0A194Q851 A0A194QQW0 A0A0L7L646 A0A3S2PLF3 H9J0A4 A0A212F0G1 H9J0A5 A0A3S2LVR9 A0A076FT24 A0A212F0F4 A0A0F6Q4J7 A0A3G1T1B2 A0A1L8D6X2 A0A212F0I1 A0A0L7KXH6 A0A0H4TG49 H9J0A8 A0A212F0G8 A0A2W1BM47 A0A0L7KNH1 A0A194QE27 A0A194QRC6 A0A0L7KNY8 A0A212F0F9 H9J0A6 A0A194Q8J5 A0A3S2NMK6 A0A194QQX1 A0A0N1IPP9 A0A194QA14 A0A2A4JLC0 U5KCB3 Q6RUR3 A0A2H1VGZ5 A0A2H1VH14 A0A140KPR8 R4FLI9 A0A194QFX2 A0A2H1VGZ4 A0A2A4JMY9 E2A713 A0A151X250 E9I9W8 A0A2A4JMW2 A0A195EUB2 A0A195B3G3 A0A195E5P2 F4W475 A0A195CJP6 A0A2H1WWP5 A0A0J7L1B2 A0A026WSD7 A0A158NDP0 E2B6N1 A0A0N0PD11 A0A158NP60 A0A1Y1N8U2 A0A067R8Z3 A0A194QA61 A0A2H1W7D8 A0A1B6DW76 A0A0N0PD56 A0A023F7C3 A0A026WK66 A0A088A4Z7 A0A224XI56 A0A1Y1MAW1 A0A2J7QR46 A0A154PC81 A0A023F7A3 A0A0P4VLL8 A0A1Y1K7W8 N6UG88 A0A0L7QSU5 A0A1W4XDZ2 A0A1W4X358 A0A1L8D6N5 J9JKN0 A0A069DY58 A0A0J7NFD4 A0A154PCJ3 A0A1Y1MP93 A0A1Y1N5U0 U5KC20 R4FM20 A0A069DQG3 A0A1W4XJR9 A0A2H8TFE7
A0A194Q851 A0A194QQW0 A0A0L7L646 A0A3S2PLF3 H9J0A4 A0A212F0G1 H9J0A5 A0A3S2LVR9 A0A076FT24 A0A212F0F4 A0A0F6Q4J7 A0A3G1T1B2 A0A1L8D6X2 A0A212F0I1 A0A0L7KXH6 A0A0H4TG49 H9J0A8 A0A212F0G8 A0A2W1BM47 A0A0L7KNH1 A0A194QE27 A0A194QRC6 A0A0L7KNY8 A0A212F0F9 H9J0A6 A0A194Q8J5 A0A3S2NMK6 A0A194QQX1 A0A0N1IPP9 A0A194QA14 A0A2A4JLC0 U5KCB3 Q6RUR3 A0A2H1VGZ5 A0A2H1VH14 A0A140KPR8 R4FLI9 A0A194QFX2 A0A2H1VGZ4 A0A2A4JMY9 E2A713 A0A151X250 E9I9W8 A0A2A4JMW2 A0A195EUB2 A0A195B3G3 A0A195E5P2 F4W475 A0A195CJP6 A0A2H1WWP5 A0A0J7L1B2 A0A026WSD7 A0A158NDP0 E2B6N1 A0A0N0PD11 A0A158NP60 A0A1Y1N8U2 A0A067R8Z3 A0A194QA61 A0A2H1W7D8 A0A1B6DW76 A0A0N0PD56 A0A023F7C3 A0A026WK66 A0A088A4Z7 A0A224XI56 A0A1Y1MAW1 A0A2J7QR46 A0A154PC81 A0A023F7A3 A0A0P4VLL8 A0A1Y1K7W8 N6UG88 A0A0L7QSU5 A0A1W4XDZ2 A0A1W4X358 A0A1L8D6N5 J9JKN0 A0A069DY58 A0A0J7NFD4 A0A154PCJ3 A0A1Y1MP93 A0A1Y1N5U0 U5KC20 R4FM20 A0A069DQG3 A0A1W4XJR9 A0A2H8TFE7
Pubmed
EMBL
BABH01019219
DQ512730
ABF59817.1
KZ149998
PZC75359.1
NWSH01000014
+ More
PCG80831.1 PCG80830.1 KP899552 AKQ06151.1 KQ459302 KPJ01717.1 KQ461175 KPJ07893.1 JTDY01002703 KOB70885.1 RSAL01000003 RVE54802.1 BABH01019201 BABH01019202 AGBW02011082 OWR47236.1 BABH01019208 RVE54801.1 KF960797 AII21999.1 OWR47235.1 KP237895 AKD01748.1 MG846919 AXY94771.1 GEYN01000012 JAV02117.1 OWR47240.1 JTDY01004799 KOB67739.1 KP899548 AKQ06147.1 BABH01019210 OWR47239.1 PZC75361.1 JTDY01007968 KOB64832.1 KPJ01716.1 KPJ07894.1 JTDY01007982 KOB64820.1 OWR47238.1 BABH01019209 KPJ01719.1 RVE54803.1 KPJ07892.1 KQ460366 KPJ15603.1 KQ459249 KPJ02363.1 NWSH01001071 PCG72771.1 KC007341 AGQ45606.1 AY491503 AAR84629.1 ODYU01002522 SOQ40120.1 SOQ40118.1 LN794625 CEO43692.1 ACPB03004191 GAHY01001368 JAA76142.1 KPJ02346.1 SOQ40119.1 PCG72772.1 GL437252 EFN70745.1 KQ982585 KYQ54300.1 GL761846 EFZ22664.1 PCG72773.1 KQ981979 KYN31469.1 KQ976625 KYM79033.1 KQ979609 KYN20169.1 GL887491 EGI71071.1 KQ977642 KYN00938.1 ODYU01011579 SOQ57407.1 LBMM01001364 KMQ96441.1 KK107119 QOIP01000009 EZA58571.1 RLU18590.1 ADTU01012731 GL446000 EFN88615.1 KPJ15584.1 ADTU01001531 GEZM01012000 GEZM01011999 GEZM01011998 GEZM01011997 GEZM01011996 GEZM01011995 GEZM01011994 GEZM01011993 JAV93300.1 KK852855 KDR14911.1 KPJ02362.1 ODYU01006680 SOQ48742.1 GEDC01007377 JAS29921.1 KPJ15601.1 GBBI01001567 JAC17145.1 KK107200 QOIP01000002 EZA55494.1 RLU25832.1 GFTR01004259 JAW12167.1 GEZM01036109 GEZM01036108 JAV82969.1 NEVH01011985 PNF31053.1 KQ434870 KZC09506.1 GBBI01001633 JAC17079.1 GDKW01001273 JAI55322.1 GEZM01089353 JAV57529.1 APGK01027734 KB740669 KB632112 ENN79646.1 ERL88916.1 KQ414755 KOC61723.1 GEYN01000013 JAV02116.1 ABLF02032246 ABLF02032258 GBGD01002580 JAC86309.1 LBMM01005734 KMQ91250.1 KZC09507.1 GEZM01028591 JAV86275.1 JAV93302.1 KC007343 AGQ45608.1 ACPB03006965 GAHY01001163 JAA76347.1 GBGD01002581 JAC86308.1 GFXV01001021 MBW12826.1
PCG80831.1 PCG80830.1 KP899552 AKQ06151.1 KQ459302 KPJ01717.1 KQ461175 KPJ07893.1 JTDY01002703 KOB70885.1 RSAL01000003 RVE54802.1 BABH01019201 BABH01019202 AGBW02011082 OWR47236.1 BABH01019208 RVE54801.1 KF960797 AII21999.1 OWR47235.1 KP237895 AKD01748.1 MG846919 AXY94771.1 GEYN01000012 JAV02117.1 OWR47240.1 JTDY01004799 KOB67739.1 KP899548 AKQ06147.1 BABH01019210 OWR47239.1 PZC75361.1 JTDY01007968 KOB64832.1 KPJ01716.1 KPJ07894.1 JTDY01007982 KOB64820.1 OWR47238.1 BABH01019209 KPJ01719.1 RVE54803.1 KPJ07892.1 KQ460366 KPJ15603.1 KQ459249 KPJ02363.1 NWSH01001071 PCG72771.1 KC007341 AGQ45606.1 AY491503 AAR84629.1 ODYU01002522 SOQ40120.1 SOQ40118.1 LN794625 CEO43692.1 ACPB03004191 GAHY01001368 JAA76142.1 KPJ02346.1 SOQ40119.1 PCG72772.1 GL437252 EFN70745.1 KQ982585 KYQ54300.1 GL761846 EFZ22664.1 PCG72773.1 KQ981979 KYN31469.1 KQ976625 KYM79033.1 KQ979609 KYN20169.1 GL887491 EGI71071.1 KQ977642 KYN00938.1 ODYU01011579 SOQ57407.1 LBMM01001364 KMQ96441.1 KK107119 QOIP01000009 EZA58571.1 RLU18590.1 ADTU01012731 GL446000 EFN88615.1 KPJ15584.1 ADTU01001531 GEZM01012000 GEZM01011999 GEZM01011998 GEZM01011997 GEZM01011996 GEZM01011995 GEZM01011994 GEZM01011993 JAV93300.1 KK852855 KDR14911.1 KPJ02362.1 ODYU01006680 SOQ48742.1 GEDC01007377 JAS29921.1 KPJ15601.1 GBBI01001567 JAC17145.1 KK107200 QOIP01000002 EZA55494.1 RLU25832.1 GFTR01004259 JAW12167.1 GEZM01036109 GEZM01036108 JAV82969.1 NEVH01011985 PNF31053.1 KQ434870 KZC09506.1 GBBI01001633 JAC17079.1 GDKW01001273 JAI55322.1 GEZM01089353 JAV57529.1 APGK01027734 KB740669 KB632112 ENN79646.1 ERL88916.1 KQ414755 KOC61723.1 GEYN01000013 JAV02116.1 ABLF02032246 ABLF02032258 GBGD01002580 JAC86309.1 LBMM01005734 KMQ91250.1 KZC09507.1 GEZM01028591 JAV86275.1 JAV93302.1 KC007343 AGQ45608.1 ACPB03006965 GAHY01001163 JAA76347.1 GBGD01002581 JAC86308.1 GFXV01001021 MBW12826.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000283053
+ More
UP000007151 UP000015103 UP000000311 UP000075809 UP000078541 UP000078540 UP000078492 UP000007755 UP000078542 UP000036403 UP000053097 UP000279307 UP000005205 UP000008237 UP000027135 UP000005203 UP000235965 UP000076502 UP000019118 UP000030742 UP000053825 UP000192223 UP000007819
UP000007151 UP000015103 UP000000311 UP000075809 UP000078541 UP000078540 UP000078492 UP000007755 UP000078542 UP000036403 UP000053097 UP000279307 UP000005205 UP000008237 UP000027135 UP000005203 UP000235965 UP000076502 UP000019118 UP000030742 UP000053825 UP000192223 UP000007819
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J0A9
Q1G151
A0A2W1BJY5
A0A2A4K9E1
A0A2A4KAR2
A0A0H4TY65
+ More
A0A194Q851 A0A194QQW0 A0A0L7L646 A0A3S2PLF3 H9J0A4 A0A212F0G1 H9J0A5 A0A3S2LVR9 A0A076FT24 A0A212F0F4 A0A0F6Q4J7 A0A3G1T1B2 A0A1L8D6X2 A0A212F0I1 A0A0L7KXH6 A0A0H4TG49 H9J0A8 A0A212F0G8 A0A2W1BM47 A0A0L7KNH1 A0A194QE27 A0A194QRC6 A0A0L7KNY8 A0A212F0F9 H9J0A6 A0A194Q8J5 A0A3S2NMK6 A0A194QQX1 A0A0N1IPP9 A0A194QA14 A0A2A4JLC0 U5KCB3 Q6RUR3 A0A2H1VGZ5 A0A2H1VH14 A0A140KPR8 R4FLI9 A0A194QFX2 A0A2H1VGZ4 A0A2A4JMY9 E2A713 A0A151X250 E9I9W8 A0A2A4JMW2 A0A195EUB2 A0A195B3G3 A0A195E5P2 F4W475 A0A195CJP6 A0A2H1WWP5 A0A0J7L1B2 A0A026WSD7 A0A158NDP0 E2B6N1 A0A0N0PD11 A0A158NP60 A0A1Y1N8U2 A0A067R8Z3 A0A194QA61 A0A2H1W7D8 A0A1B6DW76 A0A0N0PD56 A0A023F7C3 A0A026WK66 A0A088A4Z7 A0A224XI56 A0A1Y1MAW1 A0A2J7QR46 A0A154PC81 A0A023F7A3 A0A0P4VLL8 A0A1Y1K7W8 N6UG88 A0A0L7QSU5 A0A1W4XDZ2 A0A1W4X358 A0A1L8D6N5 J9JKN0 A0A069DY58 A0A0J7NFD4 A0A154PCJ3 A0A1Y1MP93 A0A1Y1N5U0 U5KC20 R4FM20 A0A069DQG3 A0A1W4XJR9 A0A2H8TFE7
A0A194Q851 A0A194QQW0 A0A0L7L646 A0A3S2PLF3 H9J0A4 A0A212F0G1 H9J0A5 A0A3S2LVR9 A0A076FT24 A0A212F0F4 A0A0F6Q4J7 A0A3G1T1B2 A0A1L8D6X2 A0A212F0I1 A0A0L7KXH6 A0A0H4TG49 H9J0A8 A0A212F0G8 A0A2W1BM47 A0A0L7KNH1 A0A194QE27 A0A194QRC6 A0A0L7KNY8 A0A212F0F9 H9J0A6 A0A194Q8J5 A0A3S2NMK6 A0A194QQX1 A0A0N1IPP9 A0A194QA14 A0A2A4JLC0 U5KCB3 Q6RUR3 A0A2H1VGZ5 A0A2H1VH14 A0A140KPR8 R4FLI9 A0A194QFX2 A0A2H1VGZ4 A0A2A4JMY9 E2A713 A0A151X250 E9I9W8 A0A2A4JMW2 A0A195EUB2 A0A195B3G3 A0A195E5P2 F4W475 A0A195CJP6 A0A2H1WWP5 A0A0J7L1B2 A0A026WSD7 A0A158NDP0 E2B6N1 A0A0N0PD11 A0A158NP60 A0A1Y1N8U2 A0A067R8Z3 A0A194QA61 A0A2H1W7D8 A0A1B6DW76 A0A0N0PD56 A0A023F7C3 A0A026WK66 A0A088A4Z7 A0A224XI56 A0A1Y1MAW1 A0A2J7QR46 A0A154PC81 A0A023F7A3 A0A0P4VLL8 A0A1Y1K7W8 N6UG88 A0A0L7QSU5 A0A1W4XDZ2 A0A1W4X358 A0A1L8D6N5 J9JKN0 A0A069DY58 A0A0J7NFD4 A0A154PCJ3 A0A1Y1MP93 A0A1Y1N5U0 U5KC20 R4FM20 A0A069DQG3 A0A1W4XJR9 A0A2H8TFE7
PDB
2GDZ
E-value=7.86537e-14,
Score=185
Ontologies
GO
Topology
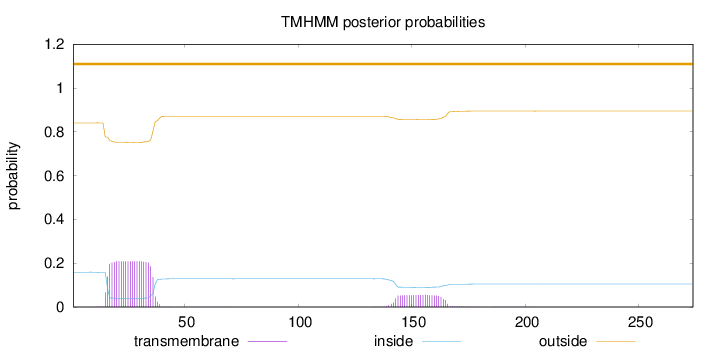
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.61028999999999
Exp number, first 60 AAs:
4.34898
Total prob of N-in:
0.15920
outside
1 - 274
Population Genetic Test Statistics
Pi
280.453263
Theta
172.450752
Tajima's D
2.747161
CLR
0.002732
CSRT
0.965051747412629
Interpretation
Uncertain
