Pre Gene Modal
BGIBMGA006895
Annotation
PREDICTED:_tetratricopeptide_repeat_protein_17-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.689 Nuclear Reliability : 1.312
Sequence
CDS
ATGTGGGACTTGCCATACTTGTCTTGTTTAGTTTTGTATATATTTTCATTTCAAGTTATTGTTATAAATTCTCGCACCTCTAATATTTGGAAATTGAATTTAGAAGAAGACCTTATTGTAGATGGCAAAACTTTGTCTCAAGATGACACAGAAAAACCATTAAGTGAGGAAGACACCGTATTCAATATAATAACATCTACAGTTTACTACGGGAATACATGGACGAAACATGGATTTGATATGTTTTGCACAGATTGTCAGATTTATGAACAACAGAGAGCTGGCAGTGAAACTAATGAAGATACACCACCATCAGAAACAAATGGGCACTCAGATGATGAATATTTAGATTGTGGCAAAGCAGTAAACTTCACTTACTATGATAACTTAGTTGGAGTTGCTAGACGACACAGACATCCTAATGTACCTGAACCACAGGTTGCATTGATATTCACAAAAAAGAAAAATGGAGTCACGGAATTTGATATAAATTCTTTAGAGAAACGTCTAAAGAAGGCGAAAAGAGAAAAACCCAAATCAGTACAGTTATACAATCAGATTGGCAACTTTTGGAGAATCAAAGGAGACACTCGACAATCGATAGAGTGTTTCCGGCGAGCACTAGCAGTGGCACCTTACAATGCTGAGGTGCTACTGAATCTGGCGCGAGTCCTGTTCACGCTGCAGTACTTGGACGACGCCATCTACCTGACGCGCCGATCGCTCGAGGTGCAGCCCCCGGACCGTAGCGCATGGCAACAGTACTTCACGCTCGGCGAGATATTCAAAGCGTACGGACATTATCAGGTACACGATTTTAATCTATACTAA
Protein
MWDLPYLSCLVLYIFSFQVIVINSRTSNIWKLNLEEDLIVDGKTLSQDDTEKPLSEEDTVFNIITSTVYYGNTWTKHGFDMFCTDCQIYEQQRAGSETNEDTPPSETNGHSDDEYLDCGKAVNFTYYDNLVGVARRHRHPNVPEPQVALIFTKKKNGVTEFDINSLEKRLKKAKREKPKSVQLYNQIGNFWRIKGDTRQSIECFRRALAVAPYNAEVLLNLARVLFTLQYLDDAIYLTRRSLEVQPPDRSAWQQYFTLGEIFKAYGHYQVHDFNLY
Summary
Uniprot
H9JBK0
A0A1E1WCU9
A0A2A4JZK5
A0A2H1WF45
A0A212ETP4
L7X3T5
+ More
A0A194Q8K0 A0A2J7PJJ0 A0A2P8XQ71 A0A067RB78 A0A1B6D116 A0A1B6DAN3 E0VG00 A0A1B6HU72 A0A1B6EXJ5 A0A224XEH6 A0A069DYS8 T1I3J5 A0A2R7W7F2 A0A1Y1LW23 D6X1D3 A0A139WBW0 A0A0A9Z1X8 A0A1W4W2L2 A0A1S4G986 A7USE5 A0A182JZ39 A0A182PFV6 A0A084WFH1 A0A182F9W4 A0A182M3E6 A0A2M4BQA6 A0A2M4BQL0 W5J840 A0A2M4BQI1 A0A182MZJ5 A0A182Y7R7 A0A182XDZ6 A0A182V1R8 A0A2M4AQ78 A0A182QJM7 A0A182UJJ3 A0A182HGM7 A0A182RH77 A0A182IKB8 F4WUY4 E2BXA7 A0A026WT09 A0A336LH52 A0A3L8DHI5 A0A151WW58 A0A195FX42 A0A195ATV6 A0A151JCJ8 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 E9IZ96 E2AAP5 A0A0L7QNJ1 A0A154PMC5 K7ITE6 A0A195CXJ4 A0A158P152 A0A0K8UVK7 A0A1D2NMT1 A0A1S3HIQ7 A0A1S3HJM9 A0A034WJL8 R7UPC8 A0A182W4I1 K1QRJ8 J3JWN5 A0A1Q3G0D8 A0A1S3D830 A0A210Q3V2 A0A2T7PPV4 A0A182T7Y3 C3XPH2 A0A0N0U2N3 A0A0K2TQ94 W4XHP6 W8B737 A0A0A9Z443 B0WE02
A0A194Q8K0 A0A2J7PJJ0 A0A2P8XQ71 A0A067RB78 A0A1B6D116 A0A1B6DAN3 E0VG00 A0A1B6HU72 A0A1B6EXJ5 A0A224XEH6 A0A069DYS8 T1I3J5 A0A2R7W7F2 A0A1Y1LW23 D6X1D3 A0A139WBW0 A0A0A9Z1X8 A0A1W4W2L2 A0A1S4G986 A7USE5 A0A182JZ39 A0A182PFV6 A0A084WFH1 A0A182F9W4 A0A182M3E6 A0A2M4BQA6 A0A2M4BQL0 W5J840 A0A2M4BQI1 A0A182MZJ5 A0A182Y7R7 A0A182XDZ6 A0A182V1R8 A0A2M4AQ78 A0A182QJM7 A0A182UJJ3 A0A182HGM7 A0A182RH77 A0A182IKB8 F4WUY4 E2BXA7 A0A026WT09 A0A336LH52 A0A3L8DHI5 A0A151WW58 A0A195FX42 A0A195ATV6 A0A151JCJ8 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 E9IZ96 E2AAP5 A0A0L7QNJ1 A0A154PMC5 K7ITE6 A0A195CXJ4 A0A158P152 A0A0K8UVK7 A0A1D2NMT1 A0A1S3HIQ7 A0A1S3HJM9 A0A034WJL8 R7UPC8 A0A182W4I1 K1QRJ8 J3JWN5 A0A1Q3G0D8 A0A1S3D830 A0A210Q3V2 A0A2T7PPV4 A0A182T7Y3 C3XPH2 A0A0N0U2N3 A0A0K2TQ94 W4XHP6 W8B737 A0A0A9Z443 B0WE02
Pubmed
19121390
22118469
23674305
26354079
29403074
24845553
+ More
20566863 26334808 28004739 18362917 19820115 25401762 26823975 12364791 24438588 20920257 23761445 25244985 21719571 20798317 24508170 30249741 21282665 20075255 21347285 27289101 25348373 23254933 22992520 22516182 28812685 18563158 24495485
20566863 26334808 28004739 18362917 19820115 25401762 26823975 12364791 24438588 20920257 23761445 25244985 21719571 20798317 24508170 30249741 21282665 20075255 21347285 27289101 25348373 23254933 22992520 22516182 28812685 18563158 24495485
EMBL
BABH01024588
BABH01024589
GDQN01006232
GDQN01000969
JAT84822.1
JAT90085.1
+ More
NWSH01000367 PCG76943.1 ODYU01007906 SOQ51094.1 AGBW02012562 OWR44821.1 KC469892 AGC92655.1 KQ459302 KPJ01724.1 NEVH01024947 PNF16502.1 PYGN01001545 PSN34156.1 KK852575 KDR21003.1 GEDC01017965 JAS19333.1 GEDC01027934 GEDC01014618 GEDC01010886 JAS09364.1 JAS22680.1 JAS26412.1 DS235129 EEB12306.1 GECU01029472 JAS78234.1 GECZ01027132 JAS42637.1 GFTR01005550 JAW10876.1 GBGD01001865 JAC87024.1 ACPB03021980 KK854357 PTY15051.1 GEZM01048943 JAV76255.1 KQ971372 EFA09505.2 KYB25417.1 GBHO01004342 GBRD01000103 GDHC01018012 GDHC01011728 GDHC01005072 JAG39262.1 JAG65718.1 JAQ00617.1 JAQ06901.1 JAQ13557.1 AAAB01008846 EDO64323.2 ATLV01023349 KE525342 KFB48965.1 AXCM01002344 GGFJ01006124 MBW55265.1 GGFJ01006153 MBW55294.1 ADMH02001968 ETN60136.1 GGFJ01006123 MBW55264.1 GGFK01009551 MBW42872.1 AXCN02001283 APCN01005677 GL888378 EGI61998.1 GL451230 EFN79690.1 KK107109 EZA59137.1 UFQS01000005 UFQT01000005 SSW96945.1 SSX17332.1 QOIP01000008 RLU19904.1 KQ982691 KYQ52056.1 KQ981200 KYN45008.1 KQ976738 KYM75663.1 KQ979038 KYN23482.1 LBMM01001700 KMQ95876.1 KZ288279 PBC29673.1 JR052404 AEY61691.1 GL767121 EFZ14085.1 GL438137 EFN69495.1 KQ414851 KOC60195.1 KQ434978 KZC13009.1 AAZX01006673 KQ977141 KYN05366.1 ADTU01005773 GDHF01021939 JAI30375.1 LJIJ01000008 ODN06246.1 GAKP01004083 JAC54869.1 AMQN01001083 KB299137 ELU08379.1 JH816675 EKC24151.1 BT127653 AEE62615.1 GFDL01001788 JAV33257.1 NEDP02005113 OWF43410.1 PZQS01000002 PVD35451.1 GG666451 EEN69843.1 KQ436033 KOX67521.1 HACA01010833 CDW28194.1 AAGJ04170915 GAMC01009575 JAB96980.1 GBHO01004345 JAG39259.1 DS231903 EDS45132.1
NWSH01000367 PCG76943.1 ODYU01007906 SOQ51094.1 AGBW02012562 OWR44821.1 KC469892 AGC92655.1 KQ459302 KPJ01724.1 NEVH01024947 PNF16502.1 PYGN01001545 PSN34156.1 KK852575 KDR21003.1 GEDC01017965 JAS19333.1 GEDC01027934 GEDC01014618 GEDC01010886 JAS09364.1 JAS22680.1 JAS26412.1 DS235129 EEB12306.1 GECU01029472 JAS78234.1 GECZ01027132 JAS42637.1 GFTR01005550 JAW10876.1 GBGD01001865 JAC87024.1 ACPB03021980 KK854357 PTY15051.1 GEZM01048943 JAV76255.1 KQ971372 EFA09505.2 KYB25417.1 GBHO01004342 GBRD01000103 GDHC01018012 GDHC01011728 GDHC01005072 JAG39262.1 JAG65718.1 JAQ00617.1 JAQ06901.1 JAQ13557.1 AAAB01008846 EDO64323.2 ATLV01023349 KE525342 KFB48965.1 AXCM01002344 GGFJ01006124 MBW55265.1 GGFJ01006153 MBW55294.1 ADMH02001968 ETN60136.1 GGFJ01006123 MBW55264.1 GGFK01009551 MBW42872.1 AXCN02001283 APCN01005677 GL888378 EGI61998.1 GL451230 EFN79690.1 KK107109 EZA59137.1 UFQS01000005 UFQT01000005 SSW96945.1 SSX17332.1 QOIP01000008 RLU19904.1 KQ982691 KYQ52056.1 KQ981200 KYN45008.1 KQ976738 KYM75663.1 KQ979038 KYN23482.1 LBMM01001700 KMQ95876.1 KZ288279 PBC29673.1 JR052404 AEY61691.1 GL767121 EFZ14085.1 GL438137 EFN69495.1 KQ414851 KOC60195.1 KQ434978 KZC13009.1 AAZX01006673 KQ977141 KYN05366.1 ADTU01005773 GDHF01021939 JAI30375.1 LJIJ01000008 ODN06246.1 GAKP01004083 JAC54869.1 AMQN01001083 KB299137 ELU08379.1 JH816675 EKC24151.1 BT127653 AEE62615.1 GFDL01001788 JAV33257.1 NEDP02005113 OWF43410.1 PZQS01000002 PVD35451.1 GG666451 EEN69843.1 KQ436033 KOX67521.1 HACA01010833 CDW28194.1 AAGJ04170915 GAMC01009575 JAB96980.1 GBHO01004345 JAG39259.1 DS231903 EDS45132.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000245037
+ More
UP000027135 UP000009046 UP000015103 UP000007266 UP000192223 UP000007062 UP000075881 UP000075885 UP000030765 UP000069272 UP000075883 UP000000673 UP000075884 UP000076408 UP000076407 UP000075903 UP000075886 UP000075902 UP000075840 UP000075900 UP000075880 UP000007755 UP000008237 UP000053097 UP000279307 UP000075809 UP000078541 UP000078540 UP000078492 UP000036403 UP000242457 UP000005203 UP000000311 UP000053825 UP000076502 UP000002358 UP000078542 UP000005205 UP000094527 UP000085678 UP000014760 UP000075920 UP000005408 UP000079169 UP000242188 UP000245119 UP000075901 UP000001554 UP000053105 UP000007110 UP000002320
UP000027135 UP000009046 UP000015103 UP000007266 UP000192223 UP000007062 UP000075881 UP000075885 UP000030765 UP000069272 UP000075883 UP000000673 UP000075884 UP000076408 UP000076407 UP000075903 UP000075886 UP000075902 UP000075840 UP000075900 UP000075880 UP000007755 UP000008237 UP000053097 UP000279307 UP000075809 UP000078541 UP000078540 UP000078492 UP000036403 UP000242457 UP000005203 UP000000311 UP000053825 UP000076502 UP000002358 UP000078542 UP000005205 UP000094527 UP000085678 UP000014760 UP000075920 UP000005408 UP000079169 UP000242188 UP000245119 UP000075901 UP000001554 UP000053105 UP000007110 UP000002320
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JBK0
A0A1E1WCU9
A0A2A4JZK5
A0A2H1WF45
A0A212ETP4
L7X3T5
+ More
A0A194Q8K0 A0A2J7PJJ0 A0A2P8XQ71 A0A067RB78 A0A1B6D116 A0A1B6DAN3 E0VG00 A0A1B6HU72 A0A1B6EXJ5 A0A224XEH6 A0A069DYS8 T1I3J5 A0A2R7W7F2 A0A1Y1LW23 D6X1D3 A0A139WBW0 A0A0A9Z1X8 A0A1W4W2L2 A0A1S4G986 A7USE5 A0A182JZ39 A0A182PFV6 A0A084WFH1 A0A182F9W4 A0A182M3E6 A0A2M4BQA6 A0A2M4BQL0 W5J840 A0A2M4BQI1 A0A182MZJ5 A0A182Y7R7 A0A182XDZ6 A0A182V1R8 A0A2M4AQ78 A0A182QJM7 A0A182UJJ3 A0A182HGM7 A0A182RH77 A0A182IKB8 F4WUY4 E2BXA7 A0A026WT09 A0A336LH52 A0A3L8DHI5 A0A151WW58 A0A195FX42 A0A195ATV6 A0A151JCJ8 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 E9IZ96 E2AAP5 A0A0L7QNJ1 A0A154PMC5 K7ITE6 A0A195CXJ4 A0A158P152 A0A0K8UVK7 A0A1D2NMT1 A0A1S3HIQ7 A0A1S3HJM9 A0A034WJL8 R7UPC8 A0A182W4I1 K1QRJ8 J3JWN5 A0A1Q3G0D8 A0A1S3D830 A0A210Q3V2 A0A2T7PPV4 A0A182T7Y3 C3XPH2 A0A0N0U2N3 A0A0K2TQ94 W4XHP6 W8B737 A0A0A9Z443 B0WE02
A0A194Q8K0 A0A2J7PJJ0 A0A2P8XQ71 A0A067RB78 A0A1B6D116 A0A1B6DAN3 E0VG00 A0A1B6HU72 A0A1B6EXJ5 A0A224XEH6 A0A069DYS8 T1I3J5 A0A2R7W7F2 A0A1Y1LW23 D6X1D3 A0A139WBW0 A0A0A9Z1X8 A0A1W4W2L2 A0A1S4G986 A7USE5 A0A182JZ39 A0A182PFV6 A0A084WFH1 A0A182F9W4 A0A182M3E6 A0A2M4BQA6 A0A2M4BQL0 W5J840 A0A2M4BQI1 A0A182MZJ5 A0A182Y7R7 A0A182XDZ6 A0A182V1R8 A0A2M4AQ78 A0A182QJM7 A0A182UJJ3 A0A182HGM7 A0A182RH77 A0A182IKB8 F4WUY4 E2BXA7 A0A026WT09 A0A336LH52 A0A3L8DHI5 A0A151WW58 A0A195FX42 A0A195ATV6 A0A151JCJ8 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 E9IZ96 E2AAP5 A0A0L7QNJ1 A0A154PMC5 K7ITE6 A0A195CXJ4 A0A158P152 A0A0K8UVK7 A0A1D2NMT1 A0A1S3HIQ7 A0A1S3HJM9 A0A034WJL8 R7UPC8 A0A182W4I1 K1QRJ8 J3JWN5 A0A1Q3G0D8 A0A1S3D830 A0A210Q3V2 A0A2T7PPV4 A0A182T7Y3 C3XPH2 A0A0N0U2N3 A0A0K2TQ94 W4XHP6 W8B737 A0A0A9Z443 B0WE02
PDB
1NA0
E-value=2.20791e-05,
Score=112
Ontologies
GO
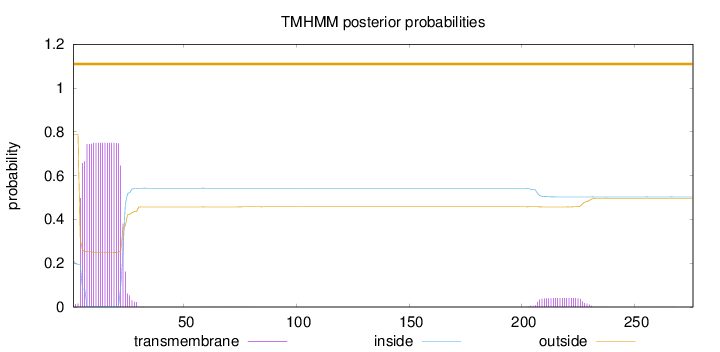
Topology
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.34656
Exp number, first 60 AAs:
14.45849
Total prob of N-in:
0.21079
POSSIBLE N-term signal
sequence
outside
1 - 276
Population Genetic Test Statistics
Pi
361.925904
Theta
220.241149
Tajima's D
2.191177
CLR
0.403003
CSRT
0.910004499775011
Interpretation
Uncertain
