Pre Gene Modal
BGIBMGA006895
Annotation
PREDICTED:_uncharacterized_protein_LOC106718385_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.136 PlasmaMembrane Reliability : 1.048
Sequence
CDS
ATGTATGGCGGAGTGTGCGTGGAAGCAGCGCTTCACTTGAGACACGCGCTGGCTCTGAAGCCGGACTTCGAGCCAGCGCTCGCCGCACTCAAAGATATCGAGAATATACCGGAGGCTTCGGTTCACGTGTACACATTACTCATCATTATTTGTTTGGTAATGGGTGTACTGCTAGTGATATTGTCGTCGGTGGACAGCGGCACCGACACGGAGGAGCCCAAGACGCAGAGACATTTTAACCGAGCGATGGCGATGCGATCTCTCCGAGGCGTCGCCTTCTCCGGCTCGAGACGGAAGAAAGGCGGATCCTAA
Protein
MYGGVCVEAALHLRHALALKPDFEPALAALKDIENIPEASVHVYTLLIIICLVMGVLLVILSSVDSGTDTEEPKTQRHFNRAMAMRSLRGVAFSGSRRKKGGS
Summary
Uniprot
A0A194QWE1
A0A1E1WCU9
A0A194Q8K0
L7X3T5
A0A212ETP4
A0A1W4W2L2
+ More
E0VG00 A0A1Y1LW23 A0A2P8XQ71 A0A1B6HU72 A0A1B6CW60 A0A1B6EXJ5 A0A1B6KH30 A0A1B6M1C5 A0A2J7PJJ0 A0A067RB78 A0A1B6DAN3 A0A139WBW0 D6X1D3 A0A182M3E6 W5J840 A0A2M4AQ78 A0A182T7Y3 A0A182Y7R7 A0A182JZ39 A0A1S4G986 A0A182V1R8 A0A182MZJ5 A0A182XDZ6 A0A182UJJ3 A0A182HGM7 A0A182PFV6 A0A182QJM7 A0A2M4BQL0 A0A2M4BQA6 A0A2M4BQI1 T1I3J5 A0A224XEH6 A0A069DYS8 A0A232EPK0 A0A0A9Z1X8 K7ITE6 A0A026WT09 A0A3L8DHI5 A0A182W4I1 E2BXA7 W8B737 A0A182RH77 T1G1U3 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 A0A034WJL8 A0A0K8UVK7 A0A182IKB8 A0A182F9W4 A0A154PMC5 A0A0N0U2N3 A0A084WFH1 E2AAP5 A0A0B7A5H3 E9IZ96 A0A0L7QNJ1 A0A2C9JEL9 F4WUY4 A0A151JCJ8 A0A195ATV6 A0A195FX42 A0A151WW58 A0A1A9UHX7 A0A195CXJ4 A0A1B0G5L8 A0A1A9Y8M7 A0A1B0BPE9 A0A1A9W940 A0A1J1HW16 A0A0L0BL46
E0VG00 A0A1Y1LW23 A0A2P8XQ71 A0A1B6HU72 A0A1B6CW60 A0A1B6EXJ5 A0A1B6KH30 A0A1B6M1C5 A0A2J7PJJ0 A0A067RB78 A0A1B6DAN3 A0A139WBW0 D6X1D3 A0A182M3E6 W5J840 A0A2M4AQ78 A0A182T7Y3 A0A182Y7R7 A0A182JZ39 A0A1S4G986 A0A182V1R8 A0A182MZJ5 A0A182XDZ6 A0A182UJJ3 A0A182HGM7 A0A182PFV6 A0A182QJM7 A0A2M4BQL0 A0A2M4BQA6 A0A2M4BQI1 T1I3J5 A0A224XEH6 A0A069DYS8 A0A232EPK0 A0A0A9Z1X8 K7ITE6 A0A026WT09 A0A3L8DHI5 A0A182W4I1 E2BXA7 W8B737 A0A182RH77 T1G1U3 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 A0A034WJL8 A0A0K8UVK7 A0A182IKB8 A0A182F9W4 A0A154PMC5 A0A0N0U2N3 A0A084WFH1 E2AAP5 A0A0B7A5H3 E9IZ96 A0A0L7QNJ1 A0A2C9JEL9 F4WUY4 A0A151JCJ8 A0A195ATV6 A0A195FX42 A0A151WW58 A0A1A9UHX7 A0A195CXJ4 A0A1B0G5L8 A0A1A9Y8M7 A0A1B0BPE9 A0A1A9W940 A0A1J1HW16 A0A0L0BL46
Pubmed
EMBL
KQ461175
KPJ07886.1
GDQN01006232
GDQN01000969
JAT84822.1
JAT90085.1
+ More
KQ459302 KPJ01724.1 KC469892 AGC92655.1 AGBW02012562 OWR44821.1 DS235129 EEB12306.1 GEZM01048943 JAV76255.1 PYGN01001545 PSN34156.1 GECU01029472 JAS78234.1 GEDC01019556 JAS17742.1 GECZ01027132 JAS42637.1 GEBQ01029513 JAT10464.1 GEBQ01010241 JAT29736.1 NEVH01024947 PNF16502.1 KK852575 KDR21003.1 GEDC01027934 GEDC01014618 GEDC01010886 JAS09364.1 JAS22680.1 JAS26412.1 KQ971372 KYB25417.1 EFA09505.2 AXCM01002344 ADMH02001968 ETN60136.1 GGFK01009551 MBW42872.1 AAAB01008846 APCN01005677 AXCN02001283 GGFJ01006153 MBW55294.1 GGFJ01006124 MBW55265.1 GGFJ01006123 MBW55264.1 ACPB03021980 GFTR01005550 JAW10876.1 GBGD01001865 JAC87024.1 NNAY01002932 OXU20273.1 GBHO01004342 GBRD01000103 GDHC01018012 GDHC01011728 GDHC01005072 JAG39262.1 JAG65718.1 JAQ00617.1 JAQ06901.1 JAQ13557.1 AAZX01006673 KK107109 EZA59137.1 QOIP01000008 RLU19904.1 GL451230 EFN79690.1 GAMC01009575 JAB96980.1 AMQM01003171 KB096023 ESO09136.1 LBMM01001700 KMQ95876.1 KZ288279 PBC29673.1 JR052404 AEY61691.1 GAKP01004083 JAC54869.1 GDHF01021939 JAI30375.1 KQ434978 KZC13009.1 KQ436033 KOX67521.1 ATLV01023349 KE525342 KFB48965.1 GL438137 EFN69495.1 HACG01029007 CEK75872.1 GL767121 EFZ14085.1 KQ414851 KOC60195.1 GL888378 EGI61998.1 KQ979038 KYN23482.1 KQ976738 KYM75663.1 KQ981200 KYN45008.1 KQ982691 KYQ52056.1 KQ977141 KYN05366.1 CCAG010013940 JXJN01017889 CVRI01000024 CRK92179.1 JRES01001701 KNC20830.1
KQ459302 KPJ01724.1 KC469892 AGC92655.1 AGBW02012562 OWR44821.1 DS235129 EEB12306.1 GEZM01048943 JAV76255.1 PYGN01001545 PSN34156.1 GECU01029472 JAS78234.1 GEDC01019556 JAS17742.1 GECZ01027132 JAS42637.1 GEBQ01029513 JAT10464.1 GEBQ01010241 JAT29736.1 NEVH01024947 PNF16502.1 KK852575 KDR21003.1 GEDC01027934 GEDC01014618 GEDC01010886 JAS09364.1 JAS22680.1 JAS26412.1 KQ971372 KYB25417.1 EFA09505.2 AXCM01002344 ADMH02001968 ETN60136.1 GGFK01009551 MBW42872.1 AAAB01008846 APCN01005677 AXCN02001283 GGFJ01006153 MBW55294.1 GGFJ01006124 MBW55265.1 GGFJ01006123 MBW55264.1 ACPB03021980 GFTR01005550 JAW10876.1 GBGD01001865 JAC87024.1 NNAY01002932 OXU20273.1 GBHO01004342 GBRD01000103 GDHC01018012 GDHC01011728 GDHC01005072 JAG39262.1 JAG65718.1 JAQ00617.1 JAQ06901.1 JAQ13557.1 AAZX01006673 KK107109 EZA59137.1 QOIP01000008 RLU19904.1 GL451230 EFN79690.1 GAMC01009575 JAB96980.1 AMQM01003171 KB096023 ESO09136.1 LBMM01001700 KMQ95876.1 KZ288279 PBC29673.1 JR052404 AEY61691.1 GAKP01004083 JAC54869.1 GDHF01021939 JAI30375.1 KQ434978 KZC13009.1 KQ436033 KOX67521.1 ATLV01023349 KE525342 KFB48965.1 GL438137 EFN69495.1 HACG01029007 CEK75872.1 GL767121 EFZ14085.1 KQ414851 KOC60195.1 GL888378 EGI61998.1 KQ979038 KYN23482.1 KQ976738 KYM75663.1 KQ981200 KYN45008.1 KQ982691 KYQ52056.1 KQ977141 KYN05366.1 CCAG010013940 JXJN01017889 CVRI01000024 CRK92179.1 JRES01001701 KNC20830.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000192223
UP000009046
UP000245037
+ More
UP000235965 UP000027135 UP000007266 UP000075883 UP000000673 UP000075901 UP000076408 UP000075881 UP000075903 UP000075884 UP000076407 UP000075902 UP000075840 UP000075885 UP000075886 UP000015103 UP000215335 UP000002358 UP000053097 UP000279307 UP000075920 UP000008237 UP000075900 UP000015101 UP000036403 UP000242457 UP000005203 UP000075880 UP000069272 UP000076502 UP000053105 UP000030765 UP000000311 UP000053825 UP000076420 UP000007755 UP000078492 UP000078540 UP000078541 UP000075809 UP000078200 UP000078542 UP000092444 UP000092443 UP000092460 UP000091820 UP000183832 UP000037069
UP000235965 UP000027135 UP000007266 UP000075883 UP000000673 UP000075901 UP000076408 UP000075881 UP000075903 UP000075884 UP000076407 UP000075902 UP000075840 UP000075885 UP000075886 UP000015103 UP000215335 UP000002358 UP000053097 UP000279307 UP000075920 UP000008237 UP000075900 UP000015101 UP000036403 UP000242457 UP000005203 UP000075880 UP000069272 UP000076502 UP000053105 UP000030765 UP000000311 UP000053825 UP000076420 UP000007755 UP000078492 UP000078540 UP000078541 UP000075809 UP000078200 UP000078542 UP000092444 UP000092443 UP000092460 UP000091820 UP000183832 UP000037069
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A194QWE1
A0A1E1WCU9
A0A194Q8K0
L7X3T5
A0A212ETP4
A0A1W4W2L2
+ More
E0VG00 A0A1Y1LW23 A0A2P8XQ71 A0A1B6HU72 A0A1B6CW60 A0A1B6EXJ5 A0A1B6KH30 A0A1B6M1C5 A0A2J7PJJ0 A0A067RB78 A0A1B6DAN3 A0A139WBW0 D6X1D3 A0A182M3E6 W5J840 A0A2M4AQ78 A0A182T7Y3 A0A182Y7R7 A0A182JZ39 A0A1S4G986 A0A182V1R8 A0A182MZJ5 A0A182XDZ6 A0A182UJJ3 A0A182HGM7 A0A182PFV6 A0A182QJM7 A0A2M4BQL0 A0A2M4BQA6 A0A2M4BQI1 T1I3J5 A0A224XEH6 A0A069DYS8 A0A232EPK0 A0A0A9Z1X8 K7ITE6 A0A026WT09 A0A3L8DHI5 A0A182W4I1 E2BXA7 W8B737 A0A182RH77 T1G1U3 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 A0A034WJL8 A0A0K8UVK7 A0A182IKB8 A0A182F9W4 A0A154PMC5 A0A0N0U2N3 A0A084WFH1 E2AAP5 A0A0B7A5H3 E9IZ96 A0A0L7QNJ1 A0A2C9JEL9 F4WUY4 A0A151JCJ8 A0A195ATV6 A0A195FX42 A0A151WW58 A0A1A9UHX7 A0A195CXJ4 A0A1B0G5L8 A0A1A9Y8M7 A0A1B0BPE9 A0A1A9W940 A0A1J1HW16 A0A0L0BL46
E0VG00 A0A1Y1LW23 A0A2P8XQ71 A0A1B6HU72 A0A1B6CW60 A0A1B6EXJ5 A0A1B6KH30 A0A1B6M1C5 A0A2J7PJJ0 A0A067RB78 A0A1B6DAN3 A0A139WBW0 D6X1D3 A0A182M3E6 W5J840 A0A2M4AQ78 A0A182T7Y3 A0A182Y7R7 A0A182JZ39 A0A1S4G986 A0A182V1R8 A0A182MZJ5 A0A182XDZ6 A0A182UJJ3 A0A182HGM7 A0A182PFV6 A0A182QJM7 A0A2M4BQL0 A0A2M4BQA6 A0A2M4BQI1 T1I3J5 A0A224XEH6 A0A069DYS8 A0A232EPK0 A0A0A9Z1X8 K7ITE6 A0A026WT09 A0A3L8DHI5 A0A182W4I1 E2BXA7 W8B737 A0A182RH77 T1G1U3 A0A0J7KZT4 A0A2A3ED17 V9IMA5 A0A088A790 A0A034WJL8 A0A0K8UVK7 A0A182IKB8 A0A182F9W4 A0A154PMC5 A0A0N0U2N3 A0A084WFH1 E2AAP5 A0A0B7A5H3 E9IZ96 A0A0L7QNJ1 A0A2C9JEL9 F4WUY4 A0A151JCJ8 A0A195ATV6 A0A195FX42 A0A151WW58 A0A1A9UHX7 A0A195CXJ4 A0A1B0G5L8 A0A1A9Y8M7 A0A1B0BPE9 A0A1A9W940 A0A1J1HW16 A0A0L0BL46
Ontologies
GO
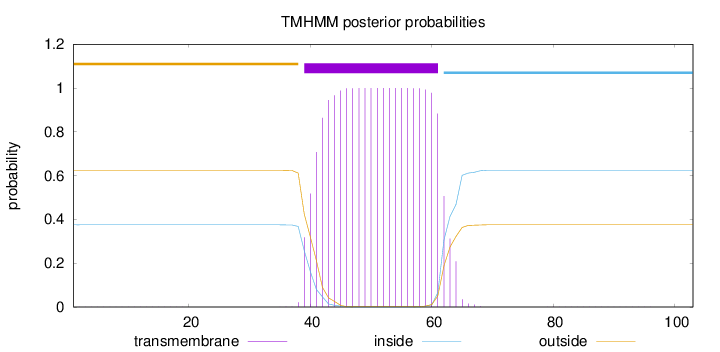
Topology
Length:
103
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.2806
Exp number, first 60 AAs:
20.3002
Total prob of N-in:
0.37558
POSSIBLE N-term signal
sequence
outside
1 - 38
TMhelix
39 - 61
inside
62 - 103
Population Genetic Test Statistics
Pi
339.541509
Theta
193.922712
Tajima's D
2.429226
CLR
0
CSRT
0.938053097345133
Interpretation
Uncertain
