Gene
KWMTBOMO05856 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006966
Annotation
PREDICTED:_probable_nuclear_transport_factor_2_isoform_X1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 2.067
Sequence
CDS
ATGGCGCTCAATCCGCAGTACGATGCAATCGGTAAAGGTTTTGTGCAACAATATTACACATTGTTCGATGATCCGGCTCAACGGGCAAATCTTGTTAATATGTACAATGTTGAAACTTCATTCATGACCTTTGAGGGAGTACAACTGCAGGGTGCTGTTAAAATTATGGAAAAATTAAATAGTTTGACTTTTCAAAAAATCACTAGAATAGTAACTGCTGTGGATTCCCAGCCAATGTTTGATGGAGGCGTTTTAATTAATGTCCTTGGAAGATTGAAGTGTGACGAAGATCCTCCACACCTATACATGCAAACGTTCGTGTTGAAGCCACTCGGAGATTCATTTTATGTTCAGCACGACATTTTCCGTTTAGGCATCCATGACATAGCTTAG
Protein
MALNPQYDAIGKGFVQQYYTLFDDPAQRANLVNMYNVETSFMTFEGVQLQGAVKIMEKLNSLTFQKITRIVTAVDSQPMFDGGVLINVLGRLKCDEDPPHLYMQTFVLKPLGDSFYVQHDIFRLGIHDIA
Summary
Uniprot
H9JBS1
A0A2A4JZW3
A0A2H1X255
I4DME9
L7X1C9
I4DJT4
+ More
B6A8J4 A0A1E1WNC8 S4PHI2 A0A1W4WE66 A0A1B6E9B0 A0A1B6MAQ7 A0A2A4JZV3 A0A194QSC6 A0A1B6F2Z3 A0A1B6HHC0 A0A1B6DXH8 D6X1D2 K7J3E1 A0A194Q9N3 A0A2A3END3 V9IJ52 A0A1S4EGD0 A0A088ACJ3 A0A067R933 A0A212ETL0 E2BIZ2 A0A2J7PTH1 A0A154PCZ3 A0A1W4WD76 A0A0L7QYW1 A0A1W4W2M9 E2AE26 A0A0N0U464 T1I158 A0A158NF22 J3JU14 A0A3L8DE56 A0A026WRC3 A0A232F9E4 R4WQ68 E0VCR2 A0A0P4VRS7 A0A0A9ZFN8 C4WRR0 A0A069DP76 R4G548 F4WEM1 A0A023F943 A0A224XYD7 A0A0L7L8A4 T1D5B7 A0A1B0DP65 A0A1B0G5L9 A0A1B0BPE3 A0A2S2N8G0 A0A2H8TJZ1 A0A1A9W936 A0A1A9Y8M5 A0A0L0BNL6 A0A0K8UIP1 A0A182Q282 A0A182PFV7 A0A182VAM7 A0A182JZ40 A0A182U0D5 A0A182XDZ5 A0A182KU75 Q7QFI2 A0A182HGM8 A0A1I8NYI3 A0A182S2A4 A0A182W4I0 A0A1B0GK96 A0A1Q3G265 B0WE01 A0A0A1WWI3 A0A0K8WHJ6 A0A182PF40 A0A0T6AWE4 A0A1J1HXW4 A0A2M3Z1J9 A0A2M4C3D6 A0A1L8EE94 A0A182MSB6 Q16UW1 A0A1I8N6X2 T1E930 A0A2M4CU11 A0A2M4AFA3 A0A182F9W5 A0A182Y291 A0A182MH47 A0A182T3N4 A0A182VJI2 A0A182NJL6 A0A182UI23 A0A182L3E5 A0A182WWQ0 Q7QIQ7
B6A8J4 A0A1E1WNC8 S4PHI2 A0A1W4WE66 A0A1B6E9B0 A0A1B6MAQ7 A0A2A4JZV3 A0A194QSC6 A0A1B6F2Z3 A0A1B6HHC0 A0A1B6DXH8 D6X1D2 K7J3E1 A0A194Q9N3 A0A2A3END3 V9IJ52 A0A1S4EGD0 A0A088ACJ3 A0A067R933 A0A212ETL0 E2BIZ2 A0A2J7PTH1 A0A154PCZ3 A0A1W4WD76 A0A0L7QYW1 A0A1W4W2M9 E2AE26 A0A0N0U464 T1I158 A0A158NF22 J3JU14 A0A3L8DE56 A0A026WRC3 A0A232F9E4 R4WQ68 E0VCR2 A0A0P4VRS7 A0A0A9ZFN8 C4WRR0 A0A069DP76 R4G548 F4WEM1 A0A023F943 A0A224XYD7 A0A0L7L8A4 T1D5B7 A0A1B0DP65 A0A1B0G5L9 A0A1B0BPE3 A0A2S2N8G0 A0A2H8TJZ1 A0A1A9W936 A0A1A9Y8M5 A0A0L0BNL6 A0A0K8UIP1 A0A182Q282 A0A182PFV7 A0A182VAM7 A0A182JZ40 A0A182U0D5 A0A182XDZ5 A0A182KU75 Q7QFI2 A0A182HGM8 A0A1I8NYI3 A0A182S2A4 A0A182W4I0 A0A1B0GK96 A0A1Q3G265 B0WE01 A0A0A1WWI3 A0A0K8WHJ6 A0A182PF40 A0A0T6AWE4 A0A1J1HXW4 A0A2M3Z1J9 A0A2M4C3D6 A0A1L8EE94 A0A182MSB6 Q16UW1 A0A1I8N6X2 T1E930 A0A2M4CU11 A0A2M4AFA3 A0A182F9W5 A0A182Y291 A0A182MH47 A0A182T3N4 A0A182VJI2 A0A182NJL6 A0A182UI23 A0A182L3E5 A0A182WWQ0 Q7QIQ7
Pubmed
19121390
22651552
23674305
23622113
26354079
18362917
+ More
19820115 20075255 24845553 22118469 20798317 21347285 22516182 30249741 24508170 28648823 23691247 20566863 27129103 25401762 26823975 26334808 21719571 25474469 26227816 24330624 26108605 20966253 12364791 14747013 17210077 25830018 17510324 25315136 25244985
19820115 20075255 24845553 22118469 20798317 21347285 22516182 30249741 24508170 28648823 23691247 20566863 27129103 25401762 26823975 26334808 21719571 25474469 26227816 24330624 26108605 20966253 12364791 14747013 17210077 25830018 17510324 25315136 25244985
EMBL
BABH01024588
NWSH01000367
PCG76942.1
ODYU01012799
SOQ59286.1
AK402467
+ More
BAM19089.1 KC469892 AGC92656.1 AK401552 BAM18174.1 DQ875254 ABK29496.2 GDQN01007751 GDQN01002559 JAT83303.1 JAT88495.1 GAIX01003267 JAA89293.1 GEDC01002788 JAS34510.1 GEBQ01006971 JAT33006.1 PCG76940.1 KQ461175 KPJ07885.1 GECZ01025199 JAS44570.1 GECU01033616 JAS74090.1 GEDC01006920 JAS30378.1 KQ971372 EFA10795.1 KQ459302 KPJ01725.1 KZ288204 PBC33227.1 JR049745 AEY61060.1 KK852620 KDR20052.1 AGBW02012562 OWR44820.1 GL448543 EFN84337.1 NEVH01021572 PNF19625.1 KQ434874 KZC09691.1 KQ414685 KOC63805.1 GL438827 EFN68259.1 KQ435840 KOX71402.1 ACPB03020977 ADTU01013709 BT126725 AEE61687.1 QOIP01000009 RLU18684.1 KK107119 EZA58528.1 NNAY01000645 OXU27222.1 AK417836 BAN21051.1 DS235063 EEB11168.1 GDKW01000823 JAI55772.1 GBHO01000788 GBRD01016800 GDHC01011662 JAG42816.1 JAG49027.1 JAQ06967.1 ABLF02034675 ABLF02034678 AK339875 BAH70580.1 GBGD01003234 JAC85655.1 GAHY01000800 JAA76710.1 GL888103 EGI67384.1 GBBI01000705 JAC18007.1 GFTR01002826 JAW13600.1 JTDY01002389 KOB71516.1 GALA01000593 JAA94259.1 AJVK01007922 CCAG010013940 CCAG010013941 JXJN01017889 GGMR01000816 MBY13435.1 GFXV01002187 MBW13992.1 JRES01001701 KNC20844.1 GDHF01025898 GDHF01024837 GDHF01012185 GDHF01008625 JAI26416.1 JAI27477.1 JAI40129.1 JAI43689.1 AXCN02000439 AAAB01008846 EAA06639.2 EGK96436.1 APCN01005678 AJWK01028576 GFDL01001140 JAV33905.1 DS231903 EDS45131.1 GBXI01011075 JAD03217.1 GDHF01024653 GDHF01001747 JAI27661.1 JAI50567.1 LJIG01022702 KRT79157.1 CVRI01000024 CRK92180.1 GGFM01001648 MBW22399.1 GGFJ01010644 MBW59785.1 GFDG01001803 JAV16996.1 AXCM01002344 CH477613 EAT38319.1 GAMD01002218 JAA99372.1 GGFL01004483 MBW68661.1 GGFK01006150 MBW39471.1 AXCM01004683 AAAB01008807 EAA04212.2
BAM19089.1 KC469892 AGC92656.1 AK401552 BAM18174.1 DQ875254 ABK29496.2 GDQN01007751 GDQN01002559 JAT83303.1 JAT88495.1 GAIX01003267 JAA89293.1 GEDC01002788 JAS34510.1 GEBQ01006971 JAT33006.1 PCG76940.1 KQ461175 KPJ07885.1 GECZ01025199 JAS44570.1 GECU01033616 JAS74090.1 GEDC01006920 JAS30378.1 KQ971372 EFA10795.1 KQ459302 KPJ01725.1 KZ288204 PBC33227.1 JR049745 AEY61060.1 KK852620 KDR20052.1 AGBW02012562 OWR44820.1 GL448543 EFN84337.1 NEVH01021572 PNF19625.1 KQ434874 KZC09691.1 KQ414685 KOC63805.1 GL438827 EFN68259.1 KQ435840 KOX71402.1 ACPB03020977 ADTU01013709 BT126725 AEE61687.1 QOIP01000009 RLU18684.1 KK107119 EZA58528.1 NNAY01000645 OXU27222.1 AK417836 BAN21051.1 DS235063 EEB11168.1 GDKW01000823 JAI55772.1 GBHO01000788 GBRD01016800 GDHC01011662 JAG42816.1 JAG49027.1 JAQ06967.1 ABLF02034675 ABLF02034678 AK339875 BAH70580.1 GBGD01003234 JAC85655.1 GAHY01000800 JAA76710.1 GL888103 EGI67384.1 GBBI01000705 JAC18007.1 GFTR01002826 JAW13600.1 JTDY01002389 KOB71516.1 GALA01000593 JAA94259.1 AJVK01007922 CCAG010013940 CCAG010013941 JXJN01017889 GGMR01000816 MBY13435.1 GFXV01002187 MBW13992.1 JRES01001701 KNC20844.1 GDHF01025898 GDHF01024837 GDHF01012185 GDHF01008625 JAI26416.1 JAI27477.1 JAI40129.1 JAI43689.1 AXCN02000439 AAAB01008846 EAA06639.2 EGK96436.1 APCN01005678 AJWK01028576 GFDL01001140 JAV33905.1 DS231903 EDS45131.1 GBXI01011075 JAD03217.1 GDHF01024653 GDHF01001747 JAI27661.1 JAI50567.1 LJIG01022702 KRT79157.1 CVRI01000024 CRK92180.1 GGFM01001648 MBW22399.1 GGFJ01010644 MBW59785.1 GFDG01001803 JAV16996.1 AXCM01002344 CH477613 EAT38319.1 GAMD01002218 JAA99372.1 GGFL01004483 MBW68661.1 GGFK01006150 MBW39471.1 AXCM01004683 AAAB01008807 EAA04212.2
Proteomes
UP000005204
UP000218220
UP000192223
UP000053240
UP000007266
UP000002358
+ More
UP000053268 UP000242457 UP000079169 UP000005203 UP000027135 UP000007151 UP000008237 UP000235965 UP000076502 UP000053825 UP000000311 UP000053105 UP000015103 UP000005205 UP000279307 UP000053097 UP000215335 UP000009046 UP000007819 UP000007755 UP000037510 UP000092462 UP000092444 UP000092460 UP000091820 UP000092443 UP000037069 UP000075886 UP000075885 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000095300 UP000075900 UP000075920 UP000092461 UP000002320 UP000183832 UP000075883 UP000008820 UP000095301 UP000069272 UP000076408 UP000075901 UP000075884
UP000053268 UP000242457 UP000079169 UP000005203 UP000027135 UP000007151 UP000008237 UP000235965 UP000076502 UP000053825 UP000000311 UP000053105 UP000015103 UP000005205 UP000279307 UP000053097 UP000215335 UP000009046 UP000007819 UP000007755 UP000037510 UP000092462 UP000092444 UP000092460 UP000091820 UP000092443 UP000037069 UP000075886 UP000075885 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000095300 UP000075900 UP000075920 UP000092461 UP000002320 UP000183832 UP000075883 UP000008820 UP000095301 UP000069272 UP000076408 UP000075901 UP000075884
PRIDE
Pfam
PF02136 NTF2
SUPFAM
SSF54427
SSF54427
CDD
ProteinModelPortal
H9JBS1
A0A2A4JZW3
A0A2H1X255
I4DME9
L7X1C9
I4DJT4
+ More
B6A8J4 A0A1E1WNC8 S4PHI2 A0A1W4WE66 A0A1B6E9B0 A0A1B6MAQ7 A0A2A4JZV3 A0A194QSC6 A0A1B6F2Z3 A0A1B6HHC0 A0A1B6DXH8 D6X1D2 K7J3E1 A0A194Q9N3 A0A2A3END3 V9IJ52 A0A1S4EGD0 A0A088ACJ3 A0A067R933 A0A212ETL0 E2BIZ2 A0A2J7PTH1 A0A154PCZ3 A0A1W4WD76 A0A0L7QYW1 A0A1W4W2M9 E2AE26 A0A0N0U464 T1I158 A0A158NF22 J3JU14 A0A3L8DE56 A0A026WRC3 A0A232F9E4 R4WQ68 E0VCR2 A0A0P4VRS7 A0A0A9ZFN8 C4WRR0 A0A069DP76 R4G548 F4WEM1 A0A023F943 A0A224XYD7 A0A0L7L8A4 T1D5B7 A0A1B0DP65 A0A1B0G5L9 A0A1B0BPE3 A0A2S2N8G0 A0A2H8TJZ1 A0A1A9W936 A0A1A9Y8M5 A0A0L0BNL6 A0A0K8UIP1 A0A182Q282 A0A182PFV7 A0A182VAM7 A0A182JZ40 A0A182U0D5 A0A182XDZ5 A0A182KU75 Q7QFI2 A0A182HGM8 A0A1I8NYI3 A0A182S2A4 A0A182W4I0 A0A1B0GK96 A0A1Q3G265 B0WE01 A0A0A1WWI3 A0A0K8WHJ6 A0A182PF40 A0A0T6AWE4 A0A1J1HXW4 A0A2M3Z1J9 A0A2M4C3D6 A0A1L8EE94 A0A182MSB6 Q16UW1 A0A1I8N6X2 T1E930 A0A2M4CU11 A0A2M4AFA3 A0A182F9W5 A0A182Y291 A0A182MH47 A0A182T3N4 A0A182VJI2 A0A182NJL6 A0A182UI23 A0A182L3E5 A0A182WWQ0 Q7QIQ7
B6A8J4 A0A1E1WNC8 S4PHI2 A0A1W4WE66 A0A1B6E9B0 A0A1B6MAQ7 A0A2A4JZV3 A0A194QSC6 A0A1B6F2Z3 A0A1B6HHC0 A0A1B6DXH8 D6X1D2 K7J3E1 A0A194Q9N3 A0A2A3END3 V9IJ52 A0A1S4EGD0 A0A088ACJ3 A0A067R933 A0A212ETL0 E2BIZ2 A0A2J7PTH1 A0A154PCZ3 A0A1W4WD76 A0A0L7QYW1 A0A1W4W2M9 E2AE26 A0A0N0U464 T1I158 A0A158NF22 J3JU14 A0A3L8DE56 A0A026WRC3 A0A232F9E4 R4WQ68 E0VCR2 A0A0P4VRS7 A0A0A9ZFN8 C4WRR0 A0A069DP76 R4G548 F4WEM1 A0A023F943 A0A224XYD7 A0A0L7L8A4 T1D5B7 A0A1B0DP65 A0A1B0G5L9 A0A1B0BPE3 A0A2S2N8G0 A0A2H8TJZ1 A0A1A9W936 A0A1A9Y8M5 A0A0L0BNL6 A0A0K8UIP1 A0A182Q282 A0A182PFV7 A0A182VAM7 A0A182JZ40 A0A182U0D5 A0A182XDZ5 A0A182KU75 Q7QFI2 A0A182HGM8 A0A1I8NYI3 A0A182S2A4 A0A182W4I0 A0A1B0GK96 A0A1Q3G265 B0WE01 A0A0A1WWI3 A0A0K8WHJ6 A0A182PF40 A0A0T6AWE4 A0A1J1HXW4 A0A2M3Z1J9 A0A2M4C3D6 A0A1L8EE94 A0A182MSB6 Q16UW1 A0A1I8N6X2 T1E930 A0A2M4CU11 A0A2M4AFA3 A0A182F9W5 A0A182Y291 A0A182MH47 A0A182T3N4 A0A182VJI2 A0A182NJL6 A0A182UI23 A0A182L3E5 A0A182WWQ0 Q7QIQ7
PDB
5BXQ
E-value=8.62026e-23,
Score=257
Ontologies
GO
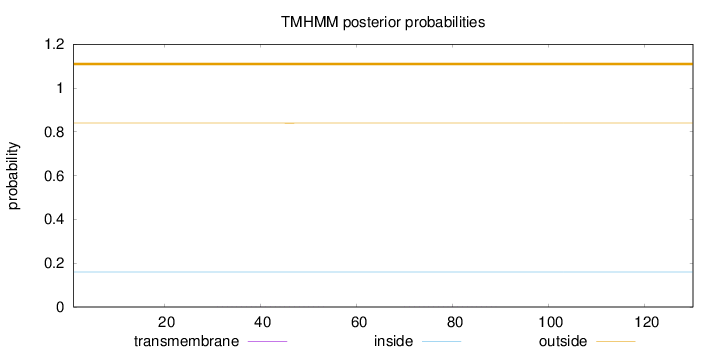
Topology
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01028
Exp number, first 60 AAs:
0.0065
Total prob of N-in:
0.15986
outside
1 - 130
Population Genetic Test Statistics
Pi
266.661533
Theta
197.976796
Tajima's D
1.173576
CLR
0.852086
CSRT
0.705714714264287
Interpretation
Uncertain
