Pre Gene Modal
BGIBMGA006896
Annotation
PREDICTED:_U4/U6_small_nuclear_ribonucleoprotein_Prp31_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.281
Sequence
CDS
ATGTCTCTAGCTGACGAATTACTTGCTGATTTAGAAGAAAATGACGATGGTGAACTCGAAGCTGCTATCGAGAGCAAAACCAAAGAAGAATACGACTTTGCAGTTCCATATCCCGTCTTACCTAAAGAAGAAGAAATCAAGAATGTCTCGATAAGAGAACTCGCGAAACTAAGATATTCTGATCGTCTGAAACGTGTTATGAGTGAAATAGAATGTAATGCTGGAAATAACAGAAATAAGGTTGAAGTTGCTGGCTTAATGGAGTCTGACCCTGAGTATCAACTGATTGTGGAAGCAAATAATGTTGCTGTTGAAATTGATGGTGAAATAGCTATTATTCACAGATTTGTCCGTGACAAATATCAGAAACGGTTTCCTGAATTGGAATCTCTAATCATCACTCCATTGGAATACATAAGGACAGTGAAAGAACTTGGCAATGATTTGGACAGGGCTAAGAATAATGAAATATTACAGAGTTTTCTTACTCAGGCCACTATTATGATTGTTTCTGTTACTGCATCTACAACTCAAGGGAAATTACTAAGCCAAATTGAACTAGAAGAAATCAATGAGGCCTGTGATATGGCAAGTGAGCTGAACAATTTCAAATCGCAAATATATGAGTATGTTGAAAGTAGAATGACTTTTATTGCACCAAACATAACTGCCATTGTTGGAGCATCAACAGCAGCAAAAATCCTTGGTGTGGCCGGTGGATTGTCTAAATTATCAAAGATGCCAGCGTGTAATGTGTTACCTCTCGGGCAACAGAAGAAGACACTGTCAGGCTTCTCGCAAGCAACAGCTCTACCTCATACTGGGTTCATTTATTTTTCACAAATTGTACAAGATACAACACCTGAATTAAGGTACAAAGCTGCGAAATTAGTGTCGACTAAGTTAACATTAGCCGCCCGTGTGGACGCGTGCCACGAGAGCACCGATGGGCACATCGGTCGGCAACTACGGGAAAATATCGAAAAGAAACTAGATAAACTACAGGAACCCCCACCGGTGAAGTTCGTGAAACCCCTACCGAAGCCGATCGAACAAAGCAGGAAGAAGAGAGGCGGCAAACGTGTCAGGAAGATGAAGGAGCGTTACGCTATGACCGAGTTCAGGAAGAACGCCAACCGACTAAACTTTGCTGACATCGAAGATGACGCGTACCAAGAAGATTTAGGATACACGCGAGGCACGATCGGCAAGTCTAGCACGGGCCGAATCCGACTACCGCAAATCGACGAGAAAACGAAGGTCCGCATCAGCAAGACATTGCAAAAGAACTTACAGAAACAACAGCAGTACGGCGGTGCTACCAGTATTAGAAGACAAGTATCTGGTACTGCGTCTTCTGTGGCCTTCACGCCTCTACAGGGTTTGGAAATCGTCAATCCTCAAGCCGCAGAAACTAGGACCAACGAAGGCAATGCCAAGTATTTCTCTAATACATCCGGGTTCTTATCTGTGGGAAAGAAATCTGATTAA
Protein
MSLADELLADLEENDDGELEAAIESKTKEEYDFAVPYPVLPKEEEIKNVSIRELAKLRYSDRLKRVMSEIECNAGNNRNKVEVAGLMESDPEYQLIVEANNVAVEIDGEIAIIHRFVRDKYQKRFPELESLIITPLEYIRTVKELGNDLDRAKNNEILQSFLTQATIMIVSVTASTTQGKLLSQIELEEINEACDMASELNNFKSQIYEYVESRMTFIAPNITAIVGASTAAKILGVAGGLSKLSKMPACNVLPLGQQKKTLSGFSQATALPHTGFIYFSQIVQDTTPELRYKAAKLVSTKLTLAARVDACHESTDGHIGRQLRENIEKKLDKLQEPPPVKFVKPLPKPIEQSRKKRGGKRVRKMKERYAMTEFRKNANRLNFADIEDDAYQEDLGYTRGTIGKSSTGRIRLPQIDEKTKVRISKTLQKNLQKQQQYGGATSIRRQVSGTASSVAFTPLQGLEIVNPQAAETRTNEGNAKYFSNTSGFLSVGKKSD
Summary
Uniprot
A0A212ETL8
A0A194QE36
A0A3S2TTI3
A0A2A4JYP9
S4PX48
A0A1E1WRZ0
+ More
A0A194QRB4 L7WW56 A0A151IWX7 A0A151I376 A0A2J7RJ76 A0A067RFN2 A0A158NMD6 A0A2P8XXZ8 E2BA59 A0A154P8B9 A0A151JWN5 F4X5S9 A0A195D781 A0A151WVF6 A0A088A4A7 A0A0J7KN77 E2AHQ5 A0A0C9QMR6 A0A0L7QT99 A0A0T6B6P1 A0A1Y1LVH0 A0A310SFF8 J3JZ24 A0A2A3E7I8 D6X2L5 A0A026W0B4 A0A1Q3EYX1 A0A182V3A0 A0A182U9M8 A0A232ELL6 A0A182P1J8 A0A182KT25 A0A182HPQ4 A0A182JV09 A0A182LRG4 K7J9U0 A0A1B6BWS1 A0A182H4M4 A0A182GSV5 A0A182WD77 J9HZ33 A0A182R1W9 B3M728 A0A182MXD9 A0A182QUI0 Q9VUM1 A0A1W4VPB8 B4PCD9 B3NI01 B4QKL1 B4HI60 A0A1J1IQD6 A0A2M4BKT2 A0A2M4BKQ1 W5JCV3 Q7PV98 B4L132 A0A182JDF3 A0A084WM29 B0W564 B4LGU8 A0A0L0C0F3 A0A1B6KA95 A0A2S2QVV7 W8BUN6 A0A182FV96 A0A0M8ZUJ3 A0A1A9WE15 A0A1S4E7W3 D3TM87 A0A1A9YEF7 A0A1B0B7X5 A0A0K8TWW6 A0A1W4XL50 B4N517 J9JPX3 A0A1I8PNC7 B4IZG0 Q2M1A7 B4GUD9 A0A023F5A4 A0A0V0GAS8 A0A0P4VR95 T1HUF0 A0A1I8MIY3 A0A069DU29 A0A087T6S9 A0A1B0FJN1 A0A1A9ZS45 E0W0J5 A0A1A9V472 A0A1L8DFQ0 A0A0A9W543 T1ISX8
A0A194QRB4 L7WW56 A0A151IWX7 A0A151I376 A0A2J7RJ76 A0A067RFN2 A0A158NMD6 A0A2P8XXZ8 E2BA59 A0A154P8B9 A0A151JWN5 F4X5S9 A0A195D781 A0A151WVF6 A0A088A4A7 A0A0J7KN77 E2AHQ5 A0A0C9QMR6 A0A0L7QT99 A0A0T6B6P1 A0A1Y1LVH0 A0A310SFF8 J3JZ24 A0A2A3E7I8 D6X2L5 A0A026W0B4 A0A1Q3EYX1 A0A182V3A0 A0A182U9M8 A0A232ELL6 A0A182P1J8 A0A182KT25 A0A182HPQ4 A0A182JV09 A0A182LRG4 K7J9U0 A0A1B6BWS1 A0A182H4M4 A0A182GSV5 A0A182WD77 J9HZ33 A0A182R1W9 B3M728 A0A182MXD9 A0A182QUI0 Q9VUM1 A0A1W4VPB8 B4PCD9 B3NI01 B4QKL1 B4HI60 A0A1J1IQD6 A0A2M4BKT2 A0A2M4BKQ1 W5JCV3 Q7PV98 B4L132 A0A182JDF3 A0A084WM29 B0W564 B4LGU8 A0A0L0C0F3 A0A1B6KA95 A0A2S2QVV7 W8BUN6 A0A182FV96 A0A0M8ZUJ3 A0A1A9WE15 A0A1S4E7W3 D3TM87 A0A1A9YEF7 A0A1B0B7X5 A0A0K8TWW6 A0A1W4XL50 B4N517 J9JPX3 A0A1I8PNC7 B4IZG0 Q2M1A7 B4GUD9 A0A023F5A4 A0A0V0GAS8 A0A0P4VR95 T1HUF0 A0A1I8MIY3 A0A069DU29 A0A087T6S9 A0A1B0FJN1 A0A1A9ZS45 E0W0J5 A0A1A9V472 A0A1L8DFQ0 A0A0A9W543 T1ISX8
Pubmed
22118469
26354079
23622113
23674305
24845553
21347285
+ More
29403074 20798317 21719571 28004739 22516182 23537049 18362917 19820115 24508170 30249741 28648823 20966253 20075255 26483478 17510324 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 12364791 24438588 26108605 24495485 20353571 15632085 25474469 27129103 25315136 26334808 20566863 25401762 26823975
29403074 20798317 21719571 28004739 22516182 23537049 18362917 19820115 24508170 30249741 28648823 20966253 20075255 26483478 17510324 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 12364791 24438588 26108605 24495485 20353571 15632085 25474469 27129103 25315136 26334808 20566863 25401762 26823975
EMBL
AGBW02012562
OWR44819.1
KQ459302
KPJ01726.1
RSAL01000003
RVE54812.1
+ More
NWSH01000367 PCG76939.1 GAIX01005663 JAA86897.1 GDQN01001265 JAT89789.1 KQ461175 KPJ07884.1 KC469892 AGC92657.1 KQ980838 KYN12339.1 KQ976507 KYM82800.1 NEVH01002996 PNF40886.1 KK852665 KDR18987.1 ADTU01020390 PYGN01001178 PSN36890.1 GL446678 EFN87401.1 KQ434842 KZC08112.1 KQ981655 KYN38603.1 GL888742 EGI58169.1 KQ976750 KYN08718.1 KQ982701 KYQ51919.1 LBMM01005167 KMQ91767.1 GL439579 EFN67035.1 GBYB01004779 GBYB01004784 JAG74546.1 JAG74551.1 KQ414748 KOC61863.1 LJIG01009468 KRT83020.1 GEZM01047337 JAV76878.1 KQ766043 OAD53756.1 APGK01043399 BT128505 KB741014 KB632204 AEE63462.1 ENN75460.1 ERL90102.1 KZ288344 PBC27675.1 KQ971372 EFA09428.1 KK107519 QOIP01000009 EZA49365.1 RLU18626.1 GFDL01014556 JAV20489.1 NNAY01003528 OXU19250.1 APCN01003267 AXCM01004908 AAZX01012697 GEDC01031561 GEDC01003034 JAS05737.1 JAS34264.1 JXUM01109816 KQ565356 KXJ71059.1 JXUM01085391 KQ563500 KXJ73729.1 CH477506 EJY57745.1 CH902618 EDV40893.1 AXCN02001696 AE014296 AY051928 AAF49655.1 AAK93352.1 CM000159 EDW94849.1 CH954178 EDV52087.1 CM000363 CM002912 EDX10522.1 KMY99715.1 CH480815 EDW41556.1 CVRI01000056 CRL01756.1 GGFJ01004432 MBW53573.1 GGFJ01004433 MBW53574.1 ADMH02001525 ETN62182.1 AAAB01008986 EAA00197.5 CH933809 EDW18189.1 ATLV01024365 KE525351 KFB51273.1 DS231841 EDS34811.1 CH940647 EDW70563.1 JRES01001072 KNC25736.1 GEBQ01031614 JAT08363.1 GGMS01012662 MBY81865.1 GAMC01005902 JAC00654.1 KQ435840 KOX71440.1 EZ422539 ADD18815.1 JXJN01009745 GDHF01033335 JAI18979.1 CH964101 EDW79456.1 ABLF02025474 CH916366 EDV97735.1 CH379069 EAL30668.2 CH479191 EDW26222.1 GBBI01002438 JAC16274.1 GECL01001618 JAP04506.1 GDKW01001033 JAI55562.1 ACPB03005522 GBGD01001271 JAC87618.1 KK113697 KFM60818.1 CCAG010020214 DS235860 EEB19151.1 GFDF01008806 JAV05278.1 GBHO01040052 GBRD01005888 GDHC01015691 GDHC01000709 JAG03552.1 JAG59933.1 JAQ02938.1 JAQ17920.1 JH431449
NWSH01000367 PCG76939.1 GAIX01005663 JAA86897.1 GDQN01001265 JAT89789.1 KQ461175 KPJ07884.1 KC469892 AGC92657.1 KQ980838 KYN12339.1 KQ976507 KYM82800.1 NEVH01002996 PNF40886.1 KK852665 KDR18987.1 ADTU01020390 PYGN01001178 PSN36890.1 GL446678 EFN87401.1 KQ434842 KZC08112.1 KQ981655 KYN38603.1 GL888742 EGI58169.1 KQ976750 KYN08718.1 KQ982701 KYQ51919.1 LBMM01005167 KMQ91767.1 GL439579 EFN67035.1 GBYB01004779 GBYB01004784 JAG74546.1 JAG74551.1 KQ414748 KOC61863.1 LJIG01009468 KRT83020.1 GEZM01047337 JAV76878.1 KQ766043 OAD53756.1 APGK01043399 BT128505 KB741014 KB632204 AEE63462.1 ENN75460.1 ERL90102.1 KZ288344 PBC27675.1 KQ971372 EFA09428.1 KK107519 QOIP01000009 EZA49365.1 RLU18626.1 GFDL01014556 JAV20489.1 NNAY01003528 OXU19250.1 APCN01003267 AXCM01004908 AAZX01012697 GEDC01031561 GEDC01003034 JAS05737.1 JAS34264.1 JXUM01109816 KQ565356 KXJ71059.1 JXUM01085391 KQ563500 KXJ73729.1 CH477506 EJY57745.1 CH902618 EDV40893.1 AXCN02001696 AE014296 AY051928 AAF49655.1 AAK93352.1 CM000159 EDW94849.1 CH954178 EDV52087.1 CM000363 CM002912 EDX10522.1 KMY99715.1 CH480815 EDW41556.1 CVRI01000056 CRL01756.1 GGFJ01004432 MBW53573.1 GGFJ01004433 MBW53574.1 ADMH02001525 ETN62182.1 AAAB01008986 EAA00197.5 CH933809 EDW18189.1 ATLV01024365 KE525351 KFB51273.1 DS231841 EDS34811.1 CH940647 EDW70563.1 JRES01001072 KNC25736.1 GEBQ01031614 JAT08363.1 GGMS01012662 MBY81865.1 GAMC01005902 JAC00654.1 KQ435840 KOX71440.1 EZ422539 ADD18815.1 JXJN01009745 GDHF01033335 JAI18979.1 CH964101 EDW79456.1 ABLF02025474 CH916366 EDV97735.1 CH379069 EAL30668.2 CH479191 EDW26222.1 GBBI01002438 JAC16274.1 GECL01001618 JAP04506.1 GDKW01001033 JAI55562.1 ACPB03005522 GBGD01001271 JAC87618.1 KK113697 KFM60818.1 CCAG010020214 DS235860 EEB19151.1 GFDF01008806 JAV05278.1 GBHO01040052 GBRD01005888 GDHC01015691 GDHC01000709 JAG03552.1 JAG59933.1 JAQ02938.1 JAQ17920.1 JH431449
Proteomes
UP000007151
UP000053268
UP000283053
UP000218220
UP000053240
UP000078492
+ More
UP000078540 UP000235965 UP000027135 UP000005205 UP000245037 UP000008237 UP000076502 UP000078541 UP000007755 UP000078542 UP000075809 UP000005203 UP000036403 UP000000311 UP000053825 UP000019118 UP000030742 UP000242457 UP000007266 UP000053097 UP000279307 UP000075903 UP000075902 UP000215335 UP000075885 UP000075882 UP000075840 UP000075881 UP000075883 UP000002358 UP000069940 UP000249989 UP000075920 UP000008820 UP000075900 UP000007801 UP000075884 UP000075886 UP000000803 UP000192221 UP000002282 UP000008711 UP000000304 UP000001292 UP000183832 UP000000673 UP000007062 UP000009192 UP000075880 UP000030765 UP000002320 UP000008792 UP000037069 UP000069272 UP000053105 UP000091820 UP000079169 UP000092443 UP000092460 UP000192223 UP000007798 UP000007819 UP000095300 UP000001070 UP000001819 UP000008744 UP000015103 UP000095301 UP000054359 UP000092444 UP000092445 UP000009046 UP000078200
UP000078540 UP000235965 UP000027135 UP000005205 UP000245037 UP000008237 UP000076502 UP000078541 UP000007755 UP000078542 UP000075809 UP000005203 UP000036403 UP000000311 UP000053825 UP000019118 UP000030742 UP000242457 UP000007266 UP000053097 UP000279307 UP000075903 UP000075902 UP000215335 UP000075885 UP000075882 UP000075840 UP000075881 UP000075883 UP000002358 UP000069940 UP000249989 UP000075920 UP000008820 UP000075900 UP000007801 UP000075884 UP000075886 UP000000803 UP000192221 UP000002282 UP000008711 UP000000304 UP000001292 UP000183832 UP000000673 UP000007062 UP000009192 UP000075880 UP000030765 UP000002320 UP000008792 UP000037069 UP000069272 UP000053105 UP000091820 UP000079169 UP000092443 UP000092460 UP000192223 UP000007798 UP000007819 UP000095300 UP000001070 UP000001819 UP000008744 UP000015103 UP000095301 UP000054359 UP000092444 UP000092445 UP000009046 UP000078200
PRIDE
Interpro
SUPFAM
SSF89124
SSF89124
Gene 3D
ProteinModelPortal
A0A212ETL8
A0A194QE36
A0A3S2TTI3
A0A2A4JYP9
S4PX48
A0A1E1WRZ0
+ More
A0A194QRB4 L7WW56 A0A151IWX7 A0A151I376 A0A2J7RJ76 A0A067RFN2 A0A158NMD6 A0A2P8XXZ8 E2BA59 A0A154P8B9 A0A151JWN5 F4X5S9 A0A195D781 A0A151WVF6 A0A088A4A7 A0A0J7KN77 E2AHQ5 A0A0C9QMR6 A0A0L7QT99 A0A0T6B6P1 A0A1Y1LVH0 A0A310SFF8 J3JZ24 A0A2A3E7I8 D6X2L5 A0A026W0B4 A0A1Q3EYX1 A0A182V3A0 A0A182U9M8 A0A232ELL6 A0A182P1J8 A0A182KT25 A0A182HPQ4 A0A182JV09 A0A182LRG4 K7J9U0 A0A1B6BWS1 A0A182H4M4 A0A182GSV5 A0A182WD77 J9HZ33 A0A182R1W9 B3M728 A0A182MXD9 A0A182QUI0 Q9VUM1 A0A1W4VPB8 B4PCD9 B3NI01 B4QKL1 B4HI60 A0A1J1IQD6 A0A2M4BKT2 A0A2M4BKQ1 W5JCV3 Q7PV98 B4L132 A0A182JDF3 A0A084WM29 B0W564 B4LGU8 A0A0L0C0F3 A0A1B6KA95 A0A2S2QVV7 W8BUN6 A0A182FV96 A0A0M8ZUJ3 A0A1A9WE15 A0A1S4E7W3 D3TM87 A0A1A9YEF7 A0A1B0B7X5 A0A0K8TWW6 A0A1W4XL50 B4N517 J9JPX3 A0A1I8PNC7 B4IZG0 Q2M1A7 B4GUD9 A0A023F5A4 A0A0V0GAS8 A0A0P4VR95 T1HUF0 A0A1I8MIY3 A0A069DU29 A0A087T6S9 A0A1B0FJN1 A0A1A9ZS45 E0W0J5 A0A1A9V472 A0A1L8DFQ0 A0A0A9W543 T1ISX8
A0A194QRB4 L7WW56 A0A151IWX7 A0A151I376 A0A2J7RJ76 A0A067RFN2 A0A158NMD6 A0A2P8XXZ8 E2BA59 A0A154P8B9 A0A151JWN5 F4X5S9 A0A195D781 A0A151WVF6 A0A088A4A7 A0A0J7KN77 E2AHQ5 A0A0C9QMR6 A0A0L7QT99 A0A0T6B6P1 A0A1Y1LVH0 A0A310SFF8 J3JZ24 A0A2A3E7I8 D6X2L5 A0A026W0B4 A0A1Q3EYX1 A0A182V3A0 A0A182U9M8 A0A232ELL6 A0A182P1J8 A0A182KT25 A0A182HPQ4 A0A182JV09 A0A182LRG4 K7J9U0 A0A1B6BWS1 A0A182H4M4 A0A182GSV5 A0A182WD77 J9HZ33 A0A182R1W9 B3M728 A0A182MXD9 A0A182QUI0 Q9VUM1 A0A1W4VPB8 B4PCD9 B3NI01 B4QKL1 B4HI60 A0A1J1IQD6 A0A2M4BKT2 A0A2M4BKQ1 W5JCV3 Q7PV98 B4L132 A0A182JDF3 A0A084WM29 B0W564 B4LGU8 A0A0L0C0F3 A0A1B6KA95 A0A2S2QVV7 W8BUN6 A0A182FV96 A0A0M8ZUJ3 A0A1A9WE15 A0A1S4E7W3 D3TM87 A0A1A9YEF7 A0A1B0B7X5 A0A0K8TWW6 A0A1W4XL50 B4N517 J9JPX3 A0A1I8PNC7 B4IZG0 Q2M1A7 B4GUD9 A0A023F5A4 A0A0V0GAS8 A0A0P4VR95 T1HUF0 A0A1I8MIY3 A0A069DU29 A0A087T6S9 A0A1B0FJN1 A0A1A9ZS45 E0W0J5 A0A1A9V472 A0A1L8DFQ0 A0A0A9W543 T1ISX8
PDB
6QX9
E-value=2.26195e-127,
Score=1167
Ontologies
GO
PANTHER
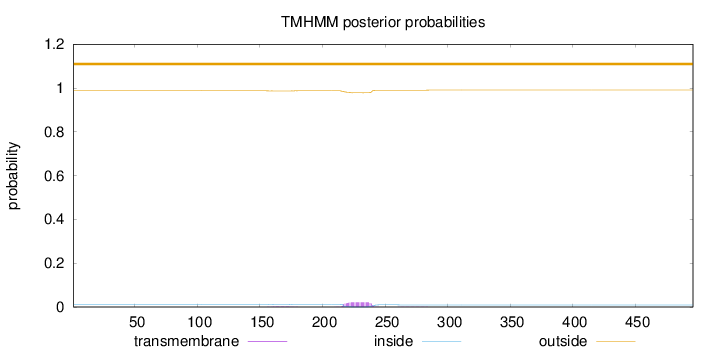
Topology
Length:
496
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.53129
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01262
outside
1 - 496
Population Genetic Test Statistics
Pi
193.277496
Theta
164.202099
Tajima's D
0.724933
CLR
0.497529
CSRT
0.578771061446928
Interpretation
Uncertain
