Gene
KWMTBOMO05852
Pre Gene Modal
BGIBMGA006898
Annotation
PREDICTED:_cerebellar_degeneration-related_protein_2-like_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.586
Sequence
CDS
ATGAGCGAGGAACAAGAACAGTGCGATTCATTCAATTCTTGGGAACTGAACGGGTTGAATTCTCTTGATCTTTGGGACTACACCGTCGAGCTGGAGTGCCTGCAAGGAACTGAAGATTTGCAACTGGCGGCCGAACTCGGGAAAACCCTACTCGAACGCAACAAGGAACTCGAGACAGCCCTTCGCCAGCATCAAAATGTTATTGAAGATCAAGCACAAGAAATCGAATATCTTACAAAGCAAACGGTTGCTTTGCGAGAGGTGAATGATTCAAGGTTGAGGATTTATGAGCAACTGGAACTCAGTATTCAAGATTTAGAACGAGCTAACCACAGACTAGCTGTCGATCATGCAGCCGACAAGAAACACATTAAGTCATTATGCTCAAACATTGACACTGTGGAGGCCAAATGCGAAGAACTACAAAAGACTGTGGACGAACTTAACGTTCAACTAGAGATCTACAGGCGAAGAGCTGAACGAAAACCCGAAAGCGAAAAACCCAAAGAGAAAGAGAAATCTGAACCATCGCAGAATTGCAAGAAACCCGAGGTAAATAGTACACCGAAGACGGTAGCACCTCTCCCGGAACTGACTAAAGAGGACGAGGATTTACTGAGGCTTAGCGATGCACTGCGAGAGAGTAAAGTGGCTTTCGCACAAGAACAGAGACGAGTCACTGAACTGGAGGAGCAATTGGCTTCTATGGTGCAAGAGAACAACAGACTACACGACCAGCTTAGGAACTGGCATACGAGAGAAGATGAGCCCAAGTCAATGCAAGAAGAATTCTCTATTCTCGAAGAAGTCAGACAAGGCCAATTGTGTATCAGGTGTCTGAGAGGTGTCGAACGAGATGACATGTCTTCTATGCTCGATGGTGATGATGACGATCGAAGCGCCATCAGTTCCGTCATATTGCCTCCATCGGGACAAGACAGCCCACGTGAAGATGTCACCAATAAACTCATCAAATTCGACAACGCCAAGGAAAAACTACTGAAAGGTACATGGGCCAATAAAGACGATGGTAATGCAAACCCATACAGAGAGCTGGTTGAAAAGTATGAGGCCCTACTCGAAGTGCAGCTGTCACAAGTTAAGAACATAAAGAAGAACAAACAGAACATTACGCCGAATTCGCAGAACCAATCAGGACCGGTCTCGCTTCAAGATGAACTTCAAACATCAGGAGATTACAGTCAGTTCAGCATCAAAGATACCGATGAGGAGAGTGGCCATGCTGATGAAACTCAAAACAACGACAAACAACCCCAGAAACCTGAGCCAACATCACGAAAGAAAATTATTCAAACCCCGGACTTTTCCGAAGCAGAGACCTCAAGCTCCGGATTTTCGGATGAGACGAGCAACAAGGCAACGCAAACTGAACGCGAGCGACCCGGTTCGTTCTTGTGTACTATCGCAGATGGAGAGGACTACCGCTTCAGCATCTATGACGACGTCAGTCCAATGGACAGTCGATTCCGCAACAGGCCGGAATATCGAGAACTATTCAAAGAGATCTTTACAATACTTAAGAAAGCGGCCGAAAATAAAGACGACGGTGAACAACTTCCCTTACTAGATGACACAACGGCGGGAAAAGTGCCTCCCGTCACTCCGGCGACGGAGGATCCCCCGGGTAACTTCACCGATGACACTCAAAGCATTTTTTCATCTGTCATGTCTGAACAATCCATTCCAGTCTCTGATATCACCGCACCGGAAACGCCCACACTCAAAGAACCAGAGAAGGAGAAACCAGATAAAGTGAAAGTCGAACCGAAAGAAAAGGTAACTCCAGAAACGAGCAAGAAGAAAGCGGTGGGTGATAACAAAGAAAACAAACCAGTGGACGATAGCACCACCGAGATATCAAACGAACAAGAGAAGGAAAAAGAACGAGTGCTCACACCGTTGGTGCGTCAGCCGCTGGAGTACATAGCGGCGACTAGAAAGAAATCCAGACATCGCAACAGGAAACACAGTCAGGACCGACAAGGCGCCGACAGCCCGGTGTATCCTACGCCGCCCAAAGTAACTTATCAGAAATCATCGAACAAGAAGAAACGTGATTATAGGCCTATAGAAGTAAGTCCGCTCGCCAGAGCTGACACCGAATGGAACGGTACCACTTTGCAATTCTACAATAGGAATTTCAACTCCCCAACACCGAGCGTGAGTGGCCGAACTGGTAAGATCTATCAAGGCTGGAACCCCGAGACAGACACGTGGGACATAAAGCAGAGCACCGCCTCGCAGGAAATACACAAACTCAGGAAGTTGGAGCTATCCTACGCCGAGGTGCTCAGAAACGCCGATAAGACTAAAACCAGGCGAAAGAAGCATCAATAA
Protein
MSEEQEQCDSFNSWELNGLNSLDLWDYTVELECLQGTEDLQLAAELGKTLLERNKELETALRQHQNVIEDQAQEIEYLTKQTVALREVNDSRLRIYEQLELSIQDLERANHRLAVDHAADKKHIKSLCSNIDTVEAKCEELQKTVDELNVQLEIYRRRAERKPESEKPKEKEKSEPSQNCKKPEVNSTPKTVAPLPELTKEDEDLLRLSDALRESKVAFAQEQRRVTELEEQLASMVQENNRLHDQLRNWHTREDEPKSMQEEFSILEEVRQGQLCIRCLRGVERDDMSSMLDGDDDDRSAISSVILPPSGQDSPREDVTNKLIKFDNAKEKLLKGTWANKDDGNANPYRELVEKYEALLEVQLSQVKNIKKNKQNITPNSQNQSGPVSLQDELQTSGDYSQFSIKDTDEESGHADETQNNDKQPQKPEPTSRKKIIQTPDFSEAETSSSGFSDETSNKATQTERERPGSFLCTIADGEDYRFSIYDDVSPMDSRFRNRPEYRELFKEIFTILKKAAENKDDGEQLPLLDDTTAGKVPPVTPATEDPPGNFTDDTQSIFSSVMSEQSIPVSDITAPETPTLKEPEKEKPDKVKVEPKEKVTPETSKKKAVGDNKENKPVDDSTTEISNEQEKEKERVLTPLVRQPLEYIAATRKKSRHRNRKHSQDRQGADSPVYPTPPKVTYQKSSNKKKRDYRPIEVSPLARADTEWNGTTLQFYNRNFNSPTPSVSGRTGKIYQGWNPETDTWDIKQSTASQEIHKLRKLELSYAEVLRNADKTKTRRKKHQ
Summary
Uniprot
H9JBK3
A0A1E1WNF6
A0A194Q875
A0A194QQW1
A0A212ETJ6
A0A2H1W6F9
+ More
A0A1E1W5C0 A0A0L7LM95 A0A1Y1N3W0 A0A1Y1N6N0 D6X2L7 T1PAR8 A0A1B6E8F7 A0A1I8ME73 T1PN34 A0A2J7QDM2 K7J9U1 A0A182H9C3 A0A0L7QPG8 A0A1B6DP59 A0A1B6C441 A0A088A4B0 E2B9J9 A0A026WR67 A0A310SHZ8 B4LWT2 U4UCP9 B4K5N0 A0A1Y1N8I0 F4WEL6 B4N1P8 A0A1J1HYQ6 A0A0J7L174 B4JT63 A0A158NF23 A0A195C652 B3NLZ1 A0A195AVG1 B4IFE6 B4Q3G3 E2AE23 A0A151WHW2 B4P6L4 Q9VIK6 A0A1W4UII6 A0A182W5R7 V5GM34 E9IWI8 A0A195FL14 A0A1I8PI60 A0A1I8PHZ7 W5JX56 A0A1L8DBI8 A0A1I8PI56 A0A1L8DBF7 A0A0L0BX88 A0A1A9XY09 A0A1B0BTR9 A0A0P6IXF0 A0A2H8TYM7 A0A1A9UM56 A0A034VK65 A0A034VHF8 A0A1A9Z8I7 A0A0A1XMQ6 A0A1A9WA55 Q17EI5 A0A0A1X2Y7 A0A034VIJ5 A0A1Q3FVU2 A0A195E802 A0A1Q3FVU5 A0A1Q3FVX5 A0A182FJU7 A0A0A9W0K7 A0A1B0FMF3 A0A0A9VT48 A0A0A9VPG3 A0A0A9W335 A0A146LJR4 A0A146LGR2
A0A1E1W5C0 A0A0L7LM95 A0A1Y1N3W0 A0A1Y1N6N0 D6X2L7 T1PAR8 A0A1B6E8F7 A0A1I8ME73 T1PN34 A0A2J7QDM2 K7J9U1 A0A182H9C3 A0A0L7QPG8 A0A1B6DP59 A0A1B6C441 A0A088A4B0 E2B9J9 A0A026WR67 A0A310SHZ8 B4LWT2 U4UCP9 B4K5N0 A0A1Y1N8I0 F4WEL6 B4N1P8 A0A1J1HYQ6 A0A0J7L174 B4JT63 A0A158NF23 A0A195C652 B3NLZ1 A0A195AVG1 B4IFE6 B4Q3G3 E2AE23 A0A151WHW2 B4P6L4 Q9VIK6 A0A1W4UII6 A0A182W5R7 V5GM34 E9IWI8 A0A195FL14 A0A1I8PI60 A0A1I8PHZ7 W5JX56 A0A1L8DBI8 A0A1I8PI56 A0A1L8DBF7 A0A0L0BX88 A0A1A9XY09 A0A1B0BTR9 A0A0P6IXF0 A0A2H8TYM7 A0A1A9UM56 A0A034VK65 A0A034VHF8 A0A1A9Z8I7 A0A0A1XMQ6 A0A1A9WA55 Q17EI5 A0A0A1X2Y7 A0A034VIJ5 A0A1Q3FVU2 A0A195E802 A0A1Q3FVU5 A0A1Q3FVX5 A0A182FJU7 A0A0A9W0K7 A0A1B0FMF3 A0A0A9VT48 A0A0A9VPG3 A0A0A9W335 A0A146LJR4 A0A146LGR2
Pubmed
19121390
26354079
22118469
26227816
28004739
18362917
+ More
19820115 25315136 20075255 26483478 20798317 24508170 30249741 17994087 18057021 23537049 21719571 21347285 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 20920257 23761445 26108605 26999592 25348373 25830018 17510324 25401762 26823975
19820115 25315136 20075255 26483478 20798317 24508170 30249741 17994087 18057021 23537049 21719571 21347285 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 20920257 23761445 26108605 26999592 25348373 25830018 17510324 25401762 26823975
EMBL
BABH01024583
GDQN01002566
JAT88488.1
KQ459302
KPJ01728.1
KQ461175
+ More
KPJ07882.1 AGBW02012562 OWR44816.1 ODYU01006576 SOQ48536.1 GDQN01008993 JAT82061.1 JTDY01000585 KOB76567.1 GEZM01013401 JAV92591.1 GEZM01013402 JAV92590.1 KQ971372 EFA09463.2 KA645837 AFP60466.1 GEDC01003089 JAS34209.1 KA649363 AFP63992.1 NEVH01015817 PNF26679.1 AAZX01003536 JXUM01120237 JXUM01120238 JXUM01120239 JXUM01120240 KQ566373 KXJ70171.1 KQ414813 KOC60530.1 GEDC01009846 JAS27452.1 GEDC01029229 JAS08069.1 GL446556 EFN87632.1 KK107119 QOIP01000009 EZA58532.1 RLU18685.1 KQ766043 OAD53759.1 CH940650 EDW66653.1 KRF82846.1 KB632204 ERL90098.1 CH933806 EDW14067.1 GEZM01013407 GEZM01013405 JAV92586.1 GL888103 EGI67379.1 CH963925 EDW78287.1 CVRI01000036 CRK93157.1 LBMM01001395 KMQ96391.1 CH916373 EDV94953.1 ADTU01013712 KQ978287 KYM95638.1 CH954179 EDV54527.1 KQ976736 KYM76047.1 CH480833 EDW46368.1 CM000361 CM002910 EDX05644.1 KMY91191.1 GL438827 EFN68256.1 KQ983106 KYQ47407.1 CM000158 EDW90966.1 AE014134 AY051930 AAF53912.1 AAK93354.1 AAN11083.1 GALX01003322 JAB65144.1 GL766526 EFZ15066.1 KQ981490 KYN41360.1 ADMH02000093 ETN67839.1 GFDF01010292 JAV03792.1 GFDF01010293 JAV03791.1 JRES01001295 KNC23849.1 JXJN01020334 GDUN01000432 JAN95487.1 GFXV01007600 MBW19405.1 GAKP01017004 JAC41948.1 GAKP01016998 JAC41954.1 GBXI01002127 JAD12165.1 CH477282 EAT44894.1 GBXI01009182 JAD05110.1 GAKP01017003 GAKP01017001 GAKP01017000 JAC41952.1 GFDL01003314 JAV31731.1 KQ979479 KYN21333.1 GFDL01003315 JAV31730.1 GFDL01003285 JAV31760.1 GBHO01045199 GBRD01017358 JAF98404.1 JAG48469.1 CCAG010000991 GBHO01045198 JAF98405.1 GBHO01045202 JAF98401.1 GBHO01042731 GBRD01017357 JAG00873.1 JAG48470.1 GDHC01011949 JAQ06680.1 GDHC01011275 JAQ07354.1
KPJ07882.1 AGBW02012562 OWR44816.1 ODYU01006576 SOQ48536.1 GDQN01008993 JAT82061.1 JTDY01000585 KOB76567.1 GEZM01013401 JAV92591.1 GEZM01013402 JAV92590.1 KQ971372 EFA09463.2 KA645837 AFP60466.1 GEDC01003089 JAS34209.1 KA649363 AFP63992.1 NEVH01015817 PNF26679.1 AAZX01003536 JXUM01120237 JXUM01120238 JXUM01120239 JXUM01120240 KQ566373 KXJ70171.1 KQ414813 KOC60530.1 GEDC01009846 JAS27452.1 GEDC01029229 JAS08069.1 GL446556 EFN87632.1 KK107119 QOIP01000009 EZA58532.1 RLU18685.1 KQ766043 OAD53759.1 CH940650 EDW66653.1 KRF82846.1 KB632204 ERL90098.1 CH933806 EDW14067.1 GEZM01013407 GEZM01013405 JAV92586.1 GL888103 EGI67379.1 CH963925 EDW78287.1 CVRI01000036 CRK93157.1 LBMM01001395 KMQ96391.1 CH916373 EDV94953.1 ADTU01013712 KQ978287 KYM95638.1 CH954179 EDV54527.1 KQ976736 KYM76047.1 CH480833 EDW46368.1 CM000361 CM002910 EDX05644.1 KMY91191.1 GL438827 EFN68256.1 KQ983106 KYQ47407.1 CM000158 EDW90966.1 AE014134 AY051930 AAF53912.1 AAK93354.1 AAN11083.1 GALX01003322 JAB65144.1 GL766526 EFZ15066.1 KQ981490 KYN41360.1 ADMH02000093 ETN67839.1 GFDF01010292 JAV03792.1 GFDF01010293 JAV03791.1 JRES01001295 KNC23849.1 JXJN01020334 GDUN01000432 JAN95487.1 GFXV01007600 MBW19405.1 GAKP01017004 JAC41948.1 GAKP01016998 JAC41954.1 GBXI01002127 JAD12165.1 CH477282 EAT44894.1 GBXI01009182 JAD05110.1 GAKP01017003 GAKP01017001 GAKP01017000 JAC41952.1 GFDL01003314 JAV31731.1 KQ979479 KYN21333.1 GFDL01003315 JAV31730.1 GFDL01003285 JAV31760.1 GBHO01045199 GBRD01017358 JAF98404.1 JAG48469.1 CCAG010000991 GBHO01045198 JAF98405.1 GBHO01045202 JAF98401.1 GBHO01042731 GBRD01017357 JAG00873.1 JAG48470.1 GDHC01011949 JAQ06680.1 GDHC01011275 JAQ07354.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000007266
+ More
UP000095301 UP000235965 UP000002358 UP000069940 UP000249989 UP000053825 UP000005203 UP000008237 UP000053097 UP000279307 UP000008792 UP000030742 UP000009192 UP000007755 UP000007798 UP000183832 UP000036403 UP000001070 UP000005205 UP000078542 UP000008711 UP000078540 UP000001292 UP000000304 UP000000311 UP000075809 UP000002282 UP000000803 UP000192221 UP000075920 UP000078541 UP000095300 UP000000673 UP000037069 UP000092443 UP000092460 UP000078200 UP000092445 UP000091820 UP000008820 UP000078492 UP000069272 UP000092444
UP000095301 UP000235965 UP000002358 UP000069940 UP000249989 UP000053825 UP000005203 UP000008237 UP000053097 UP000279307 UP000008792 UP000030742 UP000009192 UP000007755 UP000007798 UP000183832 UP000036403 UP000001070 UP000005205 UP000078542 UP000008711 UP000078540 UP000001292 UP000000304 UP000000311 UP000075809 UP000002282 UP000000803 UP000192221 UP000075920 UP000078541 UP000095300 UP000000673 UP000037069 UP000092443 UP000092460 UP000078200 UP000092445 UP000091820 UP000008820 UP000078492 UP000069272 UP000092444
Interpro
IPR026079
CDR2
ProteinModelPortal
H9JBK3
A0A1E1WNF6
A0A194Q875
A0A194QQW1
A0A212ETJ6
A0A2H1W6F9
+ More
A0A1E1W5C0 A0A0L7LM95 A0A1Y1N3W0 A0A1Y1N6N0 D6X2L7 T1PAR8 A0A1B6E8F7 A0A1I8ME73 T1PN34 A0A2J7QDM2 K7J9U1 A0A182H9C3 A0A0L7QPG8 A0A1B6DP59 A0A1B6C441 A0A088A4B0 E2B9J9 A0A026WR67 A0A310SHZ8 B4LWT2 U4UCP9 B4K5N0 A0A1Y1N8I0 F4WEL6 B4N1P8 A0A1J1HYQ6 A0A0J7L174 B4JT63 A0A158NF23 A0A195C652 B3NLZ1 A0A195AVG1 B4IFE6 B4Q3G3 E2AE23 A0A151WHW2 B4P6L4 Q9VIK6 A0A1W4UII6 A0A182W5R7 V5GM34 E9IWI8 A0A195FL14 A0A1I8PI60 A0A1I8PHZ7 W5JX56 A0A1L8DBI8 A0A1I8PI56 A0A1L8DBF7 A0A0L0BX88 A0A1A9XY09 A0A1B0BTR9 A0A0P6IXF0 A0A2H8TYM7 A0A1A9UM56 A0A034VK65 A0A034VHF8 A0A1A9Z8I7 A0A0A1XMQ6 A0A1A9WA55 Q17EI5 A0A0A1X2Y7 A0A034VIJ5 A0A1Q3FVU2 A0A195E802 A0A1Q3FVU5 A0A1Q3FVX5 A0A182FJU7 A0A0A9W0K7 A0A1B0FMF3 A0A0A9VT48 A0A0A9VPG3 A0A0A9W335 A0A146LJR4 A0A146LGR2
A0A1E1W5C0 A0A0L7LM95 A0A1Y1N3W0 A0A1Y1N6N0 D6X2L7 T1PAR8 A0A1B6E8F7 A0A1I8ME73 T1PN34 A0A2J7QDM2 K7J9U1 A0A182H9C3 A0A0L7QPG8 A0A1B6DP59 A0A1B6C441 A0A088A4B0 E2B9J9 A0A026WR67 A0A310SHZ8 B4LWT2 U4UCP9 B4K5N0 A0A1Y1N8I0 F4WEL6 B4N1P8 A0A1J1HYQ6 A0A0J7L174 B4JT63 A0A158NF23 A0A195C652 B3NLZ1 A0A195AVG1 B4IFE6 B4Q3G3 E2AE23 A0A151WHW2 B4P6L4 Q9VIK6 A0A1W4UII6 A0A182W5R7 V5GM34 E9IWI8 A0A195FL14 A0A1I8PI60 A0A1I8PHZ7 W5JX56 A0A1L8DBI8 A0A1I8PI56 A0A1L8DBF7 A0A0L0BX88 A0A1A9XY09 A0A1B0BTR9 A0A0P6IXF0 A0A2H8TYM7 A0A1A9UM56 A0A034VK65 A0A034VHF8 A0A1A9Z8I7 A0A0A1XMQ6 A0A1A9WA55 Q17EI5 A0A0A1X2Y7 A0A034VIJ5 A0A1Q3FVU2 A0A195E802 A0A1Q3FVU5 A0A1Q3FVX5 A0A182FJU7 A0A0A9W0K7 A0A1B0FMF3 A0A0A9VT48 A0A0A9VPG3 A0A0A9W335 A0A146LJR4 A0A146LGR2
Ontologies
PANTHER
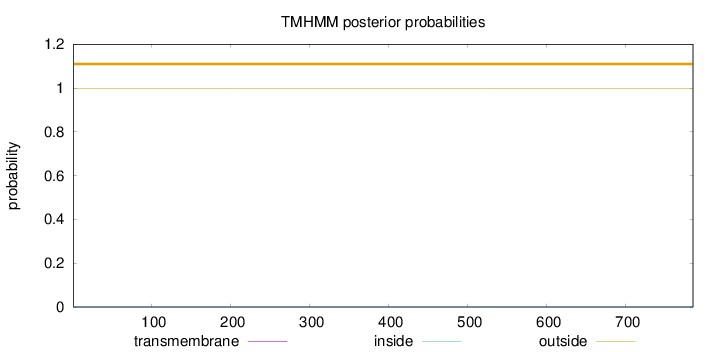
Topology
Length:
785
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00041
outside
1 - 785
Population Genetic Test Statistics
Pi
252.205605
Theta
164.673397
Tajima's D
0.312697
CLR
0.87109
CSRT
0.457227138643068
Interpretation
Uncertain
