Pre Gene Modal
BGIBMGA006964
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_F_member_3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.249 Nuclear Reliability : 2.152
Sequence
CDS
ATGGCTGGCTGTGGAGACTTCATCAAGAGTCAGTTTCCATTGATCGATGACGAATTAATGAAATATGTGGAAGATATCTTGTGCAACAGCGCTGATGAGTTTGAAGACACAGAGGAAATTTTTGAGGCCGTTGGCGAAGTTTTACAAGGCATATCTGAGAAATCTGAGAAAGACATTAAAGACATATGTGGAAAACTTCTACACATGTTACAACCTGACAAAGGATCGAAGAACAATGGGCCTACGAAAATTTTGGATGCACCAATACATTTAGCTTCAATGACCCAGACTAATTATAATGACACAGATGATCTCAAAAGTATATGGATGCAAACAAGGGATGATAGTTTGAAAGTAGATGCTAGAAAATTAGAGAAAGCCGAAGCTAAATTACAACAGAAGCAACAGAAACAAAAAGAGGTAAAAGCACCAACAGCCACACCACTATTACAAACTGCGACTGCATCACAAGTAACATCGAAAAAAGACAGCAAACTTGAAGCTAAAGGGACAAATAGAACACAAGACATAAGGATAGAAAACTTTGACATAGCGTATGGAGATCGGGTATTATTACAAGGTGCCGACTTGATACTTGCATTTGGACGAAGATACGGCTTAGTCGGAAGAAATGGTCTCGGTAAAACAACTCTCCTACGAATGATATCCAGTAAACAGTTGAAGATTCCATCGCATATAACTATCCTGCATGTGGAGCAGGAGGTTGTAGGAGATGACACCATAGCCCTGCAGAGCGTTCTTGAATGTGATACCATTAGAGAAGGCCTATTAAAAAAAGAAAGGGATATAACTGCTGCTATAAATAATGGCTCCACAGATCCGCAGCTCTCGTCCGAGTTGAGTTCAATATACGTGCAACTACAGAACATAGAGGCGGACAAGGCGCCCGCCCGGGCTTCGGTCATATTGAACGGCTTGGGTTTTACAACGGAGATGCAGGCAAGACCCACCAAGACCTTCTCCGGTGGATGGAGAATGAGATTGGCTTTGGCAAGAGCACTGTTTTCTACCCCTGACCTGTTGTTACTCGACGAACCGACCAACATGTTGGACATAAAGGCGATCATCTGGTTAGAAAACTATCTACAGAAGTGGCCCACTACCCTGTTAGTCGTCTCTCACGACAGAAATTTCTTGGACACCGTACCTACAGATATAATGCATTTACACACCCAAAGGATAGATACGTATAGGGGTAACTACGAACAGTTCCACAAGACAAAGACTGAGAAGCTGAAGAATCAGCAGCGCGAGTACGAGGCGCAGCAGCAGCACCGGGCGCACACGCAGGAGTTCATCGACCGGTTCCGTTACAACGCGAACCGCGCCTCCTCCGTGCAGAGCAAGATCAAAATGCTCGATAAACTTCCAGAACTGAAACCAGTCGAGAAGGAAATAGACGTGGTCCTGCGGTTTCCGGAGACAGAGCCGCTCTCGCCCCCCATATTGCAGCTCAACGAAGTCGGCTTCTACTATTCAAAGGACAAAGTCATATTCACAAATGTTAATCTGGGAGCGACCCTAGAATCTAGGATATGTATTGTGGGCGATAATGGCGCTGGCAAAACAACATTGCTGAAGATCATAATGGGTATACTATCGCCAACGAACGGGATCCGCACCGTGCACAGAGGGCTCAAGTTCGGTTACTTCTCGCAGCACCACGTCGATCAGCTCGAGATGAACGTCAACTCTGTTGAGTTGCTACGGAACAGTTATCCAGGTAAACCGATAGAGGAATACAGGAGGCAGTTGGGTAGTTTCGGAGTGAGCGGCGACCTCGCGTTACAGACGATAGGCAGCTTGTCCGGGGGGCAGAAGTCGAGGGTGGCCTTCGCCAGGATGTGTATGGGAAACCCTAACTTCCTAGTTCTGGACGAGCCCACCAATCATCTGGACATCGAAACGATTGAGGCTCTAGGAAAAGGAATAAACAAATACACTGGTGGCGTGATATTGGTATCTCACGACGAACGGCTAATACGGATGGTCTGCAAGGAGCTTTGGGTCTGCGGGAACGGCTGCGTCACCAGTATAGAAGGAGGATTCGACGAATATCGCAAGATAGTCGAAAGGGAATTAGAAGCACAGAATAAATAG
Protein
MAGCGDFIKSQFPLIDDELMKYVEDILCNSADEFEDTEEIFEAVGEVLQGISEKSEKDIKDICGKLLHMLQPDKGSKNNGPTKILDAPIHLASMTQTNYNDTDDLKSIWMQTRDDSLKVDARKLEKAEAKLQQKQQKQKEVKAPTATPLLQTATASQVTSKKDSKLEAKGTNRTQDIRIENFDIAYGDRVLLQGADLILAFGRRYGLVGRNGLGKTTLLRMISSKQLKIPSHITILHVEQEVVGDDTIALQSVLECDTIREGLLKKERDITAAINNGSTDPQLSSELSSIYVQLQNIEADKAPARASVILNGLGFTTEMQARPTKTFSGGWRMRLALARALFSTPDLLLLDEPTNMLDIKAIIWLENYLQKWPTTLLVVSHDRNFLDTVPTDIMHLHTQRIDTYRGNYEQFHKTKTEKLKNQQREYEAQQQHRAHTQEFIDRFRYNANRASSVQSKIKMLDKLPELKPVEKEIDVVLRFPETEPLSPPILQLNEVGFYYSKDKVIFTNVNLGATLESRICIVGDNGAGKTTLLKIIMGILSPTNGIRTVHRGLKFGYFSQHHVDQLEMNVNSVELLRNSYPGKPIEEYRRQLGSFGVSGDLALQTIGSLSGGQKSRVAFARMCMGNPNFLVLDEPTNHLDIETIEALGKGINKYTGGVILVSHDERLIRMVCKELWVCGNGCVTSIEGGFDEYRKIVERELEAQNK
Summary
Uniprot
A0A3S2LVT2
A0A2H4KAX5
H9JBR9
A0A194Q8K3
D6X393
A0A2J7PPZ8
+ More
A0A067RRH1 A0A1Y1MTU6 A0A1B6CHQ6 A0A1W4XAX0 A0A1W4X014 A0A1B6I4L3 A0A1B6LNT8 A0A1S3DDW6 J9KAX2 A0A2J7PPY7 E0V979 A0A1Q3FWK7 A0A182PKA2 Q16T85 A0A023EXN1 Q7PV55 A0A182HHT1 U5EXM4 A0A2H8TTM4 A0A182KLN4 A0A182XFR4 A0A182VK23 A0A182Q749 A0A182TW48 A0A182M0G6 A0A182JKH3 A0A182W268 A0A182JPR3 A0A182YBR2 A0A182RMR5 A0A182SU24 A0A182N9Q9 A0A084W4F5 A0A336K328 A0A0P4VZP2 A0A069DVZ0 A0A224XKP0 A0A0V0G5K9 A0A2M4A5Q7 N6T499 A0A2M3Z0C8 A0A2M3Z0K5 A0A2M3Z0J2 A0A182FU89 A0A2S2R2G1 W5JLI7 A0A146M2S6 R4FQH0 A0A2M4BF81 A0A0A9YY00 A0A0L7QSI5 A0A154PRN7 A0A310SFG8 A0A087ZST1 D1MLN5 E9IX10 A0A0C9QYH6 A0A0N0BJD4 A0A232EFN9 K7IT57 A0A0K8TMK5 A0A161MKS1 E2AC73 A0A0J7L5L9 A0A195FAW7 A0A151J261 A0A2S2QXM6 A0A151WTC5 A0A151IK01 A0A2A3EFY8 F4WWA1 A0A195B049 A0A158NKH7 E2BW60 A0A026WTT1 A0A2H4TES2 A0A034W3B8 A0A1A9UFW3 A0A0K8UHH8 A0A1B0FPV3 A0A1A9ZKM3 A0A1B0BNE6 A0A0C9RKD2 A0A1A9XY58 A0A1A9WSL8 A0A0L0BPU4 A0A0A1X9J9 A0A1S3JSX4 B3M6Z9 B4LCM1 B4IY20 T1P8S9 B4KZC3
A0A067RRH1 A0A1Y1MTU6 A0A1B6CHQ6 A0A1W4XAX0 A0A1W4X014 A0A1B6I4L3 A0A1B6LNT8 A0A1S3DDW6 J9KAX2 A0A2J7PPY7 E0V979 A0A1Q3FWK7 A0A182PKA2 Q16T85 A0A023EXN1 Q7PV55 A0A182HHT1 U5EXM4 A0A2H8TTM4 A0A182KLN4 A0A182XFR4 A0A182VK23 A0A182Q749 A0A182TW48 A0A182M0G6 A0A182JKH3 A0A182W268 A0A182JPR3 A0A182YBR2 A0A182RMR5 A0A182SU24 A0A182N9Q9 A0A084W4F5 A0A336K328 A0A0P4VZP2 A0A069DVZ0 A0A224XKP0 A0A0V0G5K9 A0A2M4A5Q7 N6T499 A0A2M3Z0C8 A0A2M3Z0K5 A0A2M3Z0J2 A0A182FU89 A0A2S2R2G1 W5JLI7 A0A146M2S6 R4FQH0 A0A2M4BF81 A0A0A9YY00 A0A0L7QSI5 A0A154PRN7 A0A310SFG8 A0A087ZST1 D1MLN5 E9IX10 A0A0C9QYH6 A0A0N0BJD4 A0A232EFN9 K7IT57 A0A0K8TMK5 A0A161MKS1 E2AC73 A0A0J7L5L9 A0A195FAW7 A0A151J261 A0A2S2QXM6 A0A151WTC5 A0A151IK01 A0A2A3EFY8 F4WWA1 A0A195B049 A0A158NKH7 E2BW60 A0A026WTT1 A0A2H4TES2 A0A034W3B8 A0A1A9UFW3 A0A0K8UHH8 A0A1B0FPV3 A0A1A9ZKM3 A0A1B0BNE6 A0A0C9RKD2 A0A1A9XY58 A0A1A9WSL8 A0A0L0BPU4 A0A0A1X9J9 A0A1S3JSX4 B3M6Z9 B4LCM1 B4IY20 T1P8S9 B4KZC3
Pubmed
19121390
26354079
18362917
19820115
24845553
28004739
+ More
20566863 17510324 24945155 26483478 12364791 14747013 17210077 20966253 25244985 24438588 27129103 26334808 23537049 20920257 23761445 26823975 25401762 20920202 21282665 28648823 20075255 26369729 20798317 21719571 21347285 24508170 30249741 29121518 25348373 26108605 25830018 17994087 25315136
20566863 17510324 24945155 26483478 12364791 14747013 17210077 20966253 25244985 24438588 27129103 26334808 23537049 20920257 23761445 26823975 25401762 20920202 21282665 28648823 20075255 26369729 20798317 21719571 21347285 24508170 30249741 29121518 25348373 26108605 25830018 17994087 25315136
EMBL
RSAL01000003
RVE54816.1
KY484786
ASS36028.1
BABH01024582
BABH01024583
+ More
KQ459302 KPJ01729.1 KQ971372 EFA09785.1 NEVH01022641 PNF18410.1 KK852510 KDR22359.1 GEZM01021182 JAV89103.1 GEDC01024329 JAS12969.1 GECU01025867 JAS81839.1 GEBQ01014681 JAT25296.1 MH172509 QAA95916.1 ABLF02027509 PNF18408.1 DS234989 EEB09935.1 GFDL01003117 JAV31928.1 CH477658 EAT37669.1 JXUM01033968 JXUM01033969 JXUM01096545 GAPW01000359 KQ564272 KQ561007 JAC13239.1 KXJ72498.1 KXJ80058.1 AAAB01008986 EAA00437.5 APCN01002438 GANO01000882 JAB58989.1 GFXV01005366 MBW17171.1 AXCN02001803 AXCM01001492 ATLV01020330 KE525298 KFB45099.1 UFQS01000062 UFQT01000062 SSW98811.1 SSX19197.1 GDKW01000544 JAI56051.1 GBGD01000641 JAC88248.1 GFTR01007825 JAW08601.1 GECL01003559 JAP02565.1 GGFK01002764 MBW36085.1 APGK01053042 APGK01053043 APGK01053044 KB741219 ENN72448.1 GGFM01001222 MBW21973.1 GGFM01001299 MBW22050.1 GGFM01001284 MBW22035.1 GGMS01014962 MBY84165.1 ADMH02000726 ETN65237.1 GDHC01005074 JAQ13555.1 GAHY01000399 JAA77111.1 GGFJ01002551 MBW51692.1 GBHO01007631 JAG35973.1 KQ414756 KOC61602.1 KQ435090 KZC14576.1 KQ760505 OAD60068.1 GU196316 ACZ26286.1 GL766616 EFZ14875.1 GBYB01005727 GBYB01007335 JAG75494.1 JAG77102.1 KQ435718 KOX78962.1 NNAY01004984 OXU17138.1 AAZX01007436 AAZX01020481 GDAI01002205 JAI15398.1 GEMB01000198 JAS02916.1 GL438428 EFN68967.1 LBMM01000640 KMQ97916.1 KQ981693 KYN37765.1 KQ980439 KYN16050.1 GGMS01013326 MBY82529.1 KQ982762 KYQ51083.1 KQ977294 KYN03962.1 KZ288266 PBC30202.1 GL888406 EGI61443.1 KQ976692 KYM77853.1 ADTU01018847 GL451091 EFN80092.1 KK107105 QOIP01000011 EZA59470.1 RLU16676.1 KY849637 ATY74518.1 GAKP01008856 JAC50096.1 GDHF01026549 GDHF01009483 JAI25765.1 JAI42831.1 CCAG010001380 JXJN01017347 GBYB01008660 JAG78427.1 JRES01001549 KNC22105.1 GBXI01006323 JAD07969.1 CH902618 EDV40864.1 CH940647 EDW69884.1 CH916366 EDV97563.1 KA645181 AFP59810.1 CH933809 EDW18949.1
KQ459302 KPJ01729.1 KQ971372 EFA09785.1 NEVH01022641 PNF18410.1 KK852510 KDR22359.1 GEZM01021182 JAV89103.1 GEDC01024329 JAS12969.1 GECU01025867 JAS81839.1 GEBQ01014681 JAT25296.1 MH172509 QAA95916.1 ABLF02027509 PNF18408.1 DS234989 EEB09935.1 GFDL01003117 JAV31928.1 CH477658 EAT37669.1 JXUM01033968 JXUM01033969 JXUM01096545 GAPW01000359 KQ564272 KQ561007 JAC13239.1 KXJ72498.1 KXJ80058.1 AAAB01008986 EAA00437.5 APCN01002438 GANO01000882 JAB58989.1 GFXV01005366 MBW17171.1 AXCN02001803 AXCM01001492 ATLV01020330 KE525298 KFB45099.1 UFQS01000062 UFQT01000062 SSW98811.1 SSX19197.1 GDKW01000544 JAI56051.1 GBGD01000641 JAC88248.1 GFTR01007825 JAW08601.1 GECL01003559 JAP02565.1 GGFK01002764 MBW36085.1 APGK01053042 APGK01053043 APGK01053044 KB741219 ENN72448.1 GGFM01001222 MBW21973.1 GGFM01001299 MBW22050.1 GGFM01001284 MBW22035.1 GGMS01014962 MBY84165.1 ADMH02000726 ETN65237.1 GDHC01005074 JAQ13555.1 GAHY01000399 JAA77111.1 GGFJ01002551 MBW51692.1 GBHO01007631 JAG35973.1 KQ414756 KOC61602.1 KQ435090 KZC14576.1 KQ760505 OAD60068.1 GU196316 ACZ26286.1 GL766616 EFZ14875.1 GBYB01005727 GBYB01007335 JAG75494.1 JAG77102.1 KQ435718 KOX78962.1 NNAY01004984 OXU17138.1 AAZX01007436 AAZX01020481 GDAI01002205 JAI15398.1 GEMB01000198 JAS02916.1 GL438428 EFN68967.1 LBMM01000640 KMQ97916.1 KQ981693 KYN37765.1 KQ980439 KYN16050.1 GGMS01013326 MBY82529.1 KQ982762 KYQ51083.1 KQ977294 KYN03962.1 KZ288266 PBC30202.1 GL888406 EGI61443.1 KQ976692 KYM77853.1 ADTU01018847 GL451091 EFN80092.1 KK107105 QOIP01000011 EZA59470.1 RLU16676.1 KY849637 ATY74518.1 GAKP01008856 JAC50096.1 GDHF01026549 GDHF01009483 JAI25765.1 JAI42831.1 CCAG010001380 JXJN01017347 GBYB01008660 JAG78427.1 JRES01001549 KNC22105.1 GBXI01006323 JAD07969.1 CH902618 EDV40864.1 CH940647 EDW69884.1 CH916366 EDV97563.1 KA645181 AFP59810.1 CH933809 EDW18949.1
Proteomes
UP000283053
UP000005204
UP000053268
UP000007266
UP000235965
UP000027135
+ More
UP000192223 UP000079169 UP000007819 UP000009046 UP000075885 UP000008820 UP000069940 UP000249989 UP000007062 UP000075840 UP000075882 UP000076407 UP000075903 UP000075886 UP000075902 UP000075883 UP000075880 UP000075920 UP000075881 UP000076408 UP000075900 UP000075901 UP000075884 UP000030765 UP000019118 UP000069272 UP000000673 UP000053825 UP000076502 UP000005203 UP000053105 UP000215335 UP000002358 UP000000311 UP000036403 UP000078541 UP000078492 UP000075809 UP000078542 UP000242457 UP000007755 UP000078540 UP000005205 UP000008237 UP000053097 UP000279307 UP000078200 UP000092444 UP000092445 UP000092460 UP000092443 UP000091820 UP000037069 UP000085678 UP000007801 UP000008792 UP000001070 UP000095301 UP000009192
UP000192223 UP000079169 UP000007819 UP000009046 UP000075885 UP000008820 UP000069940 UP000249989 UP000007062 UP000075840 UP000075882 UP000076407 UP000075903 UP000075886 UP000075902 UP000075883 UP000075880 UP000075920 UP000075881 UP000076408 UP000075900 UP000075901 UP000075884 UP000030765 UP000019118 UP000069272 UP000000673 UP000053825 UP000076502 UP000005203 UP000053105 UP000215335 UP000002358 UP000000311 UP000036403 UP000078541 UP000078492 UP000075809 UP000078542 UP000242457 UP000007755 UP000078540 UP000005205 UP000008237 UP000053097 UP000279307 UP000078200 UP000092444 UP000092445 UP000092460 UP000092443 UP000091820 UP000037069 UP000085678 UP000007801 UP000008792 UP000001070 UP000095301 UP000009192
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A3S2LVT2
A0A2H4KAX5
H9JBR9
A0A194Q8K3
D6X393
A0A2J7PPZ8
+ More
A0A067RRH1 A0A1Y1MTU6 A0A1B6CHQ6 A0A1W4XAX0 A0A1W4X014 A0A1B6I4L3 A0A1B6LNT8 A0A1S3DDW6 J9KAX2 A0A2J7PPY7 E0V979 A0A1Q3FWK7 A0A182PKA2 Q16T85 A0A023EXN1 Q7PV55 A0A182HHT1 U5EXM4 A0A2H8TTM4 A0A182KLN4 A0A182XFR4 A0A182VK23 A0A182Q749 A0A182TW48 A0A182M0G6 A0A182JKH3 A0A182W268 A0A182JPR3 A0A182YBR2 A0A182RMR5 A0A182SU24 A0A182N9Q9 A0A084W4F5 A0A336K328 A0A0P4VZP2 A0A069DVZ0 A0A224XKP0 A0A0V0G5K9 A0A2M4A5Q7 N6T499 A0A2M3Z0C8 A0A2M3Z0K5 A0A2M3Z0J2 A0A182FU89 A0A2S2R2G1 W5JLI7 A0A146M2S6 R4FQH0 A0A2M4BF81 A0A0A9YY00 A0A0L7QSI5 A0A154PRN7 A0A310SFG8 A0A087ZST1 D1MLN5 E9IX10 A0A0C9QYH6 A0A0N0BJD4 A0A232EFN9 K7IT57 A0A0K8TMK5 A0A161MKS1 E2AC73 A0A0J7L5L9 A0A195FAW7 A0A151J261 A0A2S2QXM6 A0A151WTC5 A0A151IK01 A0A2A3EFY8 F4WWA1 A0A195B049 A0A158NKH7 E2BW60 A0A026WTT1 A0A2H4TES2 A0A034W3B8 A0A1A9UFW3 A0A0K8UHH8 A0A1B0FPV3 A0A1A9ZKM3 A0A1B0BNE6 A0A0C9RKD2 A0A1A9XY58 A0A1A9WSL8 A0A0L0BPU4 A0A0A1X9J9 A0A1S3JSX4 B3M6Z9 B4LCM1 B4IY20 T1P8S9 B4KZC3
A0A067RRH1 A0A1Y1MTU6 A0A1B6CHQ6 A0A1W4XAX0 A0A1W4X014 A0A1B6I4L3 A0A1B6LNT8 A0A1S3DDW6 J9KAX2 A0A2J7PPY7 E0V979 A0A1Q3FWK7 A0A182PKA2 Q16T85 A0A023EXN1 Q7PV55 A0A182HHT1 U5EXM4 A0A2H8TTM4 A0A182KLN4 A0A182XFR4 A0A182VK23 A0A182Q749 A0A182TW48 A0A182M0G6 A0A182JKH3 A0A182W268 A0A182JPR3 A0A182YBR2 A0A182RMR5 A0A182SU24 A0A182N9Q9 A0A084W4F5 A0A336K328 A0A0P4VZP2 A0A069DVZ0 A0A224XKP0 A0A0V0G5K9 A0A2M4A5Q7 N6T499 A0A2M3Z0C8 A0A2M3Z0K5 A0A2M3Z0J2 A0A182FU89 A0A2S2R2G1 W5JLI7 A0A146M2S6 R4FQH0 A0A2M4BF81 A0A0A9YY00 A0A0L7QSI5 A0A154PRN7 A0A310SFG8 A0A087ZST1 D1MLN5 E9IX10 A0A0C9QYH6 A0A0N0BJD4 A0A232EFN9 K7IT57 A0A0K8TMK5 A0A161MKS1 E2AC73 A0A0J7L5L9 A0A195FAW7 A0A151J261 A0A2S2QXM6 A0A151WTC5 A0A151IK01 A0A2A3EFY8 F4WWA1 A0A195B049 A0A158NKH7 E2BW60 A0A026WTT1 A0A2H4TES2 A0A034W3B8 A0A1A9UFW3 A0A0K8UHH8 A0A1B0FPV3 A0A1A9ZKM3 A0A1B0BNE6 A0A0C9RKD2 A0A1A9XY58 A0A1A9WSL8 A0A0L0BPU4 A0A0A1X9J9 A0A1S3JSX4 B3M6Z9 B4LCM1 B4IY20 T1P8S9 B4KZC3
PDB
5ZXD
E-value=4.46939e-111,
Score=1028
Ontologies
GO
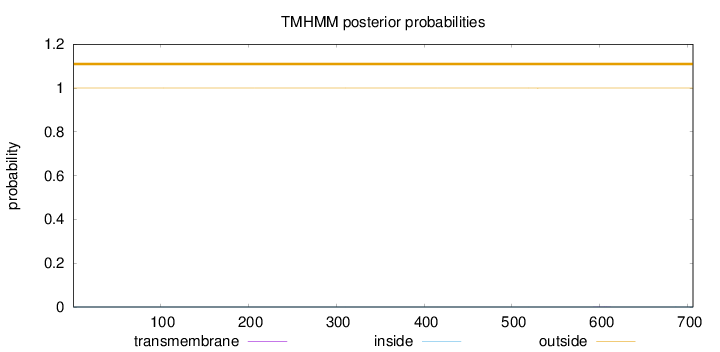
Topology
Length:
706
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00546999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00015
outside
1 - 706
Population Genetic Test Statistics
Pi
211.995435
Theta
181.251548
Tajima's D
0.48211
CLR
0.521085
CSRT
0.504774761261937
Interpretation
Uncertain
