Pre Gene Modal
BGIBMGA006959
Annotation
PREDICTED:_armadillo_segment_polarity_protein_isoform_X1_[Bombyx_mori]
Full name
Armadillo segment polarity protein
Location in the cell
Cytoplasmic Reliability : 1.708
Sequence
CDS
ATGAGTTATCAAATTCCATCATCGCAAAGTCGCACAATGTCGCACAGCAGCTATGTTGCTTCTGATGTGCCAATGGCACCAAACAAAGAGCAGCAGACATTGATGTGGCAGCAAAACTCCTATTTAGTGGACTCTGGAATCAACTCTGGAGCTGCTACCCAGGTTCCATCACTAACTGGCAAGGAAGATGATGAGATGGAAGGTGACCAACTTATGTTTGACCTTGATCAGGGTTTTGCTCAGGGATTCACTCAAGAACAAGTTGATGATATGAATCAACAGCTTTCTCAAACTAGATCTCAAAGAGTCCGTGCGGCTATGTTTCCTGAAACGCTGGAGGAAGGAATTGAAATTCCATCGACACAGCTGGATCCCGCTCAACCAACTGCTGTCCAGCGTTTAGCTGAACCTTCCCAGATGTTAAAGCATGCTGTGGTGAACCTCATCAATTATCAAGATGATGCTGATTTAGCTACCAGAGCTATTCCAGAGTTGATAAAGCTACTGAATGATGAAGATCAAGTAGTAGTATCCCAAGCTGCCATGATGGTGCATCAGCTCTCTAAGAAGGAAGCATCGCGTCACGCCATTATGAACTCGCCACAAATGGTGGCTGCGTTGGTTCGCGCAATTTCCAACAGCAATGATCTAGAAACTACTAAGGGGGCAGTTGGCACCTTACACAATTTATCTCACCATCGTCAAGGTCTGCTTGCCATCTTCAAGAGTGGTGGAATTCCAGCATTAGTGAAACTACTCAGCTCTCCAGTTGAAGCTGTGCTCTTTTATGCTATTACAACACTGCATAACCTATTGTTGCATCAAGACGGTTCAAAGATGGCTGTTCGTCTAGCTGGAGGTCTACAAAAAATGGTGGCATTACTACAAAGGAACAATGTCAAGTTCCTAGCTATTGTGACTGACTGCCTTCAGATTTTGGCCTATGGGAATCAAGAATCGAAATTGATTATTCTAGCATCTCAGGGCCCAATCGAATTAGTTCGCATTATGCGTTCTTACGATTACGAAAAGTTGCTGTGGACAACTTCGAGAGTTCTTAAGGTATTATCGGTATGCTCAAGCAACAAGCCAGCTATAGTGGAAGCTGGTGGCATGCAAGCTCTGGCCATGCATCTAGGCAACCCGAGCGGCCGTCTCGTCCAGAACTGCCTTTGGACTCTGAGGAATCTGTCTGATGCTGCTACCAAAGTGGAAGGCCTGGAGGCATTGCTGCAAAGCCTCGTGCAAGTTCTAGCTTCGACTGATGTCAACATCGTGACCTGTGCTGCTGGAATTCTCTCCAACTTGACTTGCAACAACCAACGCAATAAGATGACGGTATGCCAGGTGGGCGGCGTGGACGCGCTGGTCCGCACGGTGGTGTCGGCCGGGGACCGCGAGGAGATCACGGAGCCCGCCATCTGTGCGCTGCGACACCTCACCTCCAGGCATGACGACAGTGAGATGGCGCAGAATGCAGTACGCCTGCATTATGGATTGCCGGTGATAGTGAAGCTGCTACAGCCTCCTTCTCGCTGGCCTCTAGTGAAGGCCGTCGTGGGCTTGGTACGCAACCTGGCGCTCTGCTCCGCCAACTACGCTCCGTTGCGGGAACATGGCGCCGTTCATCATCTGGTGCGCCTGCTGATGCGTGCCTACAACGACACCCAGAGACAACGCAGCTCTTCGGGCGGTGGAGCCAATACTGCTTACGCTGACGGCGTTCGTATGGAGGAGATCGTGGAAGGCGCCGTTGGAGCCTTGCATATTTTAGCCAAAGAAAGCCACAACAGACAACTTATCCGACAGCAGAACGTCATTCCGATATTCGTGCAACTGTTGTTCAATGAGATTGAAAATATTCAGCGCGTTGCCGCCGGCGTGCTCTGTGAATTGGCCGTAGAGAAAGAAGGAGCAGAAATGATCGAAGCCGAAGGGGCAACCGCTCCGCTCACTGAATTGCTGCATTCAAGAAATGAAGGGGTCGCCACGTACGCTGCAGCCGTTCTGTTCCGCATGTCCGAAGACAAGCCCCACGATTACAAGAAGAGACTCTCTATGGAATTGACTAATTCGTTGTTCCGCGACGATCACCAGATGTGGTCCAATGACCTTCCGATACAATCCGATATACAGGACATGCTCGGGCCAGAGCAGGGTTACGAGGGCCTGTATGGGACCAGACCATCCTTTCACCAACAAGGCTACGATCAAATACCGATAGATTCGATGCAGGGGCTTGAGATCGGTAGCGGTTTTGGTATGGACATGGACATTGGCGAGGCGGAAGGCGCGGGCGCGGCGTCAGCTGACCTGGCCTTCCCCGAGCCACCCCACGACAACAACAATGTGGCAGCCTGGTACGACACAGACCTTTAA
Protein
MSYQIPSSQSRTMSHSSYVASDVPMAPNKEQQTLMWQQNSYLVDSGINSGAATQVPSLTGKEDDEMEGDQLMFDLDQGFAQGFTQEQVDDMNQQLSQTRSQRVRAAMFPETLEEGIEIPSTQLDPAQPTAVQRLAEPSQMLKHAVVNLINYQDDADLATRAIPELIKLLNDEDQVVVSQAAMMVHQLSKKEASRHAIMNSPQMVAALVRAISNSNDLETTKGAVGTLHNLSHHRQGLLAIFKSGGIPALVKLLSSPVEAVLFYAITTLHNLLLHQDGSKMAVRLAGGLQKMVALLQRNNVKFLAIVTDCLQILAYGNQESKLIILASQGPIELVRIMRSYDYEKLLWTTSRVLKVLSVCSSNKPAIVEAGGMQALAMHLGNPSGRLVQNCLWTLRNLSDAATKVEGLEALLQSLVQVLASTDVNIVTCAAGILSNLTCNNQRNKMTVCQVGGVDALVRTVVSAGDREEITEPAICALRHLTSRHDDSEMAQNAVRLHYGLPVIVKLLQPPSRWPLVKAVVGLVRNLALCSANYAPLREHGAVHHLVRLLMRAYNDTQRQRSSSGGGANTAYADGVRMEEIVEGAVGALHILAKESHNRQLIRQQNVIPIFVQLLFNEIENIQRVAAGVLCELAVEKEGAEMIEAEGATAPLTELLHSRNEGVATYAAAVLFRMSEDKPHDYKKRLSMELTNSLFRDDHQMWSNDLPIQSDIQDMLGPEQGYEGLYGTRPSFHQQGYDQIPIDSMQGLEIGSGFGMDMDIGEAEGAGAASADLAFPEPPHDNNNVAAWYDTDL
Summary
Description
Isoform neural may associate with CadN and participate in the transmission of developmental information. Can associate with alpha-catenin. Isoform cytoplasmic accumulates through wg signaling; arm function in wg signal transduction is required early in development for determination of neuroblast fate. Arm and Abl proteins function cooperatively at adherens junctions in both the CNS and epidermis.
May associate with CadN and participate in the transmission of developmental information. Can associate with alpha-catenin. Accumulates through wg signaling; arm function in wg signal transduction is required early in development for determination of neuroblast fate. Arm and Abl proteins function cooperatively at adherens junctions in both the CNS and epidermis (By similarity).
May associate with CadN and participate in the transmission of developmental information. Can associate with alpha-catenin. Accumulates through wg signaling; arm function in wg signal transduction is required early in development for determination of neuroblast fate. Arm and Abl proteins function cooperatively at adherens junctions in both the CNS and epidermis (By similarity).
Subunit
Interacts with Mer and Moe at the adherens junction (PubMed:8666669). Interacts with Inx2 (PubMed:15047872). Interacts with alpha-Cat (PubMed:25653389). Interacts with Myo31DF (PubMed:16598259, PubMed:22491943).
Interacts with Mer and Moe at the adherens junction.
Interacts with Mer and Moe at the adherens junction.
Similarity
Belongs to the beta-catenin family.
Keywords
Alternative splicing
Cell adhesion
Cell junction
Cell membrane
Complete proteome
Cytoplasm
Developmental protein
Membrane
Phosphoprotein
Reference proteome
Repeat
Segmentation polarity protein
Wnt signaling pathway
Feature
chain Armadillo segment polarity protein
splice variant In isoform Neural.
splice variant In isoform Neural.
Uniprot
H9JBR4
A0A2H1WLR9
A0A3S2NML4
X2L6G3
A0A194QE46
A0A212F6N0
+ More
A0A194QQU0 E2B7R3 A0A1L8DMB5 E2BXN9 E2AZN9 A0A0C9RQG9 A0A0L0C373 A0A0A1WGQ3 A0A026WDG5 A0A3B0KVR9 A0A1I8PYB0 A0A1I8PYE2 W8ASL5 A0A1W4VSQ9 B4M333 K7J6I5 M9PGG4 B3P8S9 P18824 A0A034VUY2 A0A0K8U2B4 B3MXH2 B4N1V6 A0A158ND49 B4JWY3 A0A3L8E4M5 A0A2A3E2T1 A0A088ASV7 A0A232EPB5 T1PFC0 A0A151X3X9 A0A195DUF5 F4X7D1 A0A195BNB7 A0A195F7X9 A0A0M4EUK6 Q02453 A0A151I8E6 A0A0M4ESV3 T1HHR2 A0A023F5L4 A0A1Y1NCG6 A0A069DWH2 A0A154PF68 A0A0T6AU15 A0A1B0DKX3 A0A0L7R6W9 A0A146L5T1 D3TLQ8 A0A336M2A0 Q29I35 Q76CZ7 A0A2J7QDQ0 D6X202 A0A1A9VD85 A0A067QTQ8 Q7QHW5 E0V8W8 A0A1B6C185 A0A2M4A3U4 A0A2M3Z1N7 A0A2M4BDL7 A0A2M4A3V6 Q17GS9 A0A2M4CUX6 A0A2M4A3R4 A0A2M4A3L0 E0V8X0 A0A182FIY5 A0A0C9RCC0 A0A0A1X9P9 A0A0K8V7Z9 A0A1I8PYE7 W8BQR6 A0A182U3Z2 A0A182P393 A0A0K8W0W9 A0A182Y567 A0A1L8DLC2 A0A088AVH5 A0A182GGV2 A0A0P6IJT4 U5EXF9 A0A1J1J1N0 A0A067QU74 A0A182RHI9 B0WHS4 A0A182W5J9 A0A084VLT8 A0A182GBL0 A0A1Q3FFC2 P18824-2 A0A1W4VS30 A0A3B0K6X4
A0A194QQU0 E2B7R3 A0A1L8DMB5 E2BXN9 E2AZN9 A0A0C9RQG9 A0A0L0C373 A0A0A1WGQ3 A0A026WDG5 A0A3B0KVR9 A0A1I8PYB0 A0A1I8PYE2 W8ASL5 A0A1W4VSQ9 B4M333 K7J6I5 M9PGG4 B3P8S9 P18824 A0A034VUY2 A0A0K8U2B4 B3MXH2 B4N1V6 A0A158ND49 B4JWY3 A0A3L8E4M5 A0A2A3E2T1 A0A088ASV7 A0A232EPB5 T1PFC0 A0A151X3X9 A0A195DUF5 F4X7D1 A0A195BNB7 A0A195F7X9 A0A0M4EUK6 Q02453 A0A151I8E6 A0A0M4ESV3 T1HHR2 A0A023F5L4 A0A1Y1NCG6 A0A069DWH2 A0A154PF68 A0A0T6AU15 A0A1B0DKX3 A0A0L7R6W9 A0A146L5T1 D3TLQ8 A0A336M2A0 Q29I35 Q76CZ7 A0A2J7QDQ0 D6X202 A0A1A9VD85 A0A067QTQ8 Q7QHW5 E0V8W8 A0A1B6C185 A0A2M4A3U4 A0A2M3Z1N7 A0A2M4BDL7 A0A2M4A3V6 Q17GS9 A0A2M4CUX6 A0A2M4A3R4 A0A2M4A3L0 E0V8X0 A0A182FIY5 A0A0C9RCC0 A0A0A1X9P9 A0A0K8V7Z9 A0A1I8PYE7 W8BQR6 A0A182U3Z2 A0A182P393 A0A0K8W0W9 A0A182Y567 A0A1L8DLC2 A0A088AVH5 A0A182GGV2 A0A0P6IJT4 U5EXF9 A0A1J1J1N0 A0A067QU74 A0A182RHI9 B0WHS4 A0A182W5J9 A0A084VLT8 A0A182GBL0 A0A1Q3FFC2 P18824-2 A0A1W4VS30 A0A3B0K6X4
Pubmed
19121390
25038464
26354079
22118469
20798317
26108605
+ More
25830018 24508170 24495485 17994087 18057021 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2707602 9635189 10731137 12537569 7529201 8666669 11927557 15047872 16598259 18327897 22491943 25653389 25348373 21347285 30249741 28648823 21719571 8483160 25474469 28004739 26334808 26823975 20353571 15632085 18362917 19820115 24845553 12364791 20566863 17510324 25244985 26483478 24438588
25830018 24508170 24495485 17994087 18057021 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2707602 9635189 10731137 12537569 7529201 8666669 11927557 15047872 16598259 18327897 22491943 25653389 25348373 21347285 30249741 28648823 21719571 8483160 25474469 28004739 26334808 26823975 20353571 15632085 18362917 19820115 24845553 12364791 20566863 17510324 25244985 26483478 24438588
EMBL
BABH01024557
BABH01024558
ODYU01009524
SOQ54020.1
RSAL01000003
RVE54822.1
+ More
KJ206237 AHN95656.1 KQ459302 KPJ01736.1 AGBW02010000 OWR49369.1 KQ461175 KPJ07873.1 GL446201 EFN88276.1 GFDF01006569 JAV07515.1 GL451281 EFN79557.1 GL444275 EFN61098.1 GBYB01010740 JAG80507.1 JRES01001065 KNC25859.1 GBXI01016225 JAC98066.1 KK107260 EZA54105.1 OUUW01000042 SPP89966.1 GAMC01014690 JAB91865.1 CH940651 EDW65208.1 KRF82078.1 KRF82079.1 AE014298 AGB94995.1 CH954183 EDV45534.1 KQS26004.1 KQS26006.1 KQS26007.1 X54468 AF001213 AL021106 AL021086 AY118525 GAKP01012733 JAC46219.1 GDHF01031839 GDHF01013604 GDHF01010197 GDHF01000012 JAI20475.1 JAI38710.1 JAI42117.1 JAI52302.1 CH902630 EDV38437.1 CH963925 EDW78345.1 KRF98857.1 ADTU01012387 CH916376 EDV95259.1 QOIP01000001 RLU27641.1 KZ288418 PBC26015.1 NNAY01002985 OXU20172.1 KA646865 AFP61494.1 KQ982554 KYQ55126.1 KQ980341 KYN16392.1 GL888828 EGI57743.1 KQ976438 KYM86857.1 KQ981744 KYN36486.1 CP012528 ALC49451.1 L04874 KQ978377 KYM94561.1 ALC49452.1 ACPB03000442 GBBI01002258 JAC16454.1 GEZM01006917 JAV95573.1 GBGD01000511 JAC88378.1 KQ434890 KZC10491.1 LJIG01022843 KRT78429.1 AJVK01006519 KQ414646 KOC66593.1 GDHC01016122 JAQ02507.1 CCAG010022197 EZ422360 ADD18636.1 UFQT01000445 SSX24365.1 CH379064 AB109212 BAD00045.1 NEVH01015817 PNF26703.1 KQ971371 EFA10737.2 KK852946 KDR13423.1 AAAB01008811 DS234986 EEB09824.1 GEDC01030303 GEDC01005515 JAS06995.1 JAS31783.1 GGFK01002071 MBW35392.1 GGFM01001675 MBW22426.1 GGFJ01002009 MBW51150.1 GGFK01002089 MBW35410.1 CH477256 EAT45858.1 GGFL01004460 MBW68638.1 GGFK01002078 MBW35399.1 GGFK01002072 MBW35393.1 EEB09826.1 GBYB01010742 JAG80509.1 GBXI01006263 JAD08029.1 GDHF01017257 GDHF01010333 GDHF01006251 JAI35057.1 JAI41981.1 JAI46063.1 GAMC01014691 JAB91864.1 GDHF01021598 GDHF01019605 GDHF01007595 JAI30716.1 JAI32709.1 JAI44719.1 GFDF01006828 JAV07256.1 JXUM01062538 JXUM01062539 JXUM01062540 JXUM01062541 JXUM01062542 KQ562204 KXJ76424.1 GDIQ01003271 JAN91466.1 GANO01000970 JAB58901.1 CVRI01000063 CRL04769.1 KDR13424.1 DS231939 EDS27941.1 ATLV01014553 ATLV01014554 KE524974 KFB38932.1 JXUM01052918 KQ561755 KXJ77622.1 GFDL01008843 JAV26202.1 SPP89967.1
KJ206237 AHN95656.1 KQ459302 KPJ01736.1 AGBW02010000 OWR49369.1 KQ461175 KPJ07873.1 GL446201 EFN88276.1 GFDF01006569 JAV07515.1 GL451281 EFN79557.1 GL444275 EFN61098.1 GBYB01010740 JAG80507.1 JRES01001065 KNC25859.1 GBXI01016225 JAC98066.1 KK107260 EZA54105.1 OUUW01000042 SPP89966.1 GAMC01014690 JAB91865.1 CH940651 EDW65208.1 KRF82078.1 KRF82079.1 AE014298 AGB94995.1 CH954183 EDV45534.1 KQS26004.1 KQS26006.1 KQS26007.1 X54468 AF001213 AL021106 AL021086 AY118525 GAKP01012733 JAC46219.1 GDHF01031839 GDHF01013604 GDHF01010197 GDHF01000012 JAI20475.1 JAI38710.1 JAI42117.1 JAI52302.1 CH902630 EDV38437.1 CH963925 EDW78345.1 KRF98857.1 ADTU01012387 CH916376 EDV95259.1 QOIP01000001 RLU27641.1 KZ288418 PBC26015.1 NNAY01002985 OXU20172.1 KA646865 AFP61494.1 KQ982554 KYQ55126.1 KQ980341 KYN16392.1 GL888828 EGI57743.1 KQ976438 KYM86857.1 KQ981744 KYN36486.1 CP012528 ALC49451.1 L04874 KQ978377 KYM94561.1 ALC49452.1 ACPB03000442 GBBI01002258 JAC16454.1 GEZM01006917 JAV95573.1 GBGD01000511 JAC88378.1 KQ434890 KZC10491.1 LJIG01022843 KRT78429.1 AJVK01006519 KQ414646 KOC66593.1 GDHC01016122 JAQ02507.1 CCAG010022197 EZ422360 ADD18636.1 UFQT01000445 SSX24365.1 CH379064 AB109212 BAD00045.1 NEVH01015817 PNF26703.1 KQ971371 EFA10737.2 KK852946 KDR13423.1 AAAB01008811 DS234986 EEB09824.1 GEDC01030303 GEDC01005515 JAS06995.1 JAS31783.1 GGFK01002071 MBW35392.1 GGFM01001675 MBW22426.1 GGFJ01002009 MBW51150.1 GGFK01002089 MBW35410.1 CH477256 EAT45858.1 GGFL01004460 MBW68638.1 GGFK01002078 MBW35399.1 GGFK01002072 MBW35393.1 EEB09826.1 GBYB01010742 JAG80509.1 GBXI01006263 JAD08029.1 GDHF01017257 GDHF01010333 GDHF01006251 JAI35057.1 JAI41981.1 JAI46063.1 GAMC01014691 JAB91864.1 GDHF01021598 GDHF01019605 GDHF01007595 JAI30716.1 JAI32709.1 JAI44719.1 GFDF01006828 JAV07256.1 JXUM01062538 JXUM01062539 JXUM01062540 JXUM01062541 JXUM01062542 KQ562204 KXJ76424.1 GDIQ01003271 JAN91466.1 GANO01000970 JAB58901.1 CVRI01000063 CRL04769.1 KDR13424.1 DS231939 EDS27941.1 ATLV01014553 ATLV01014554 KE524974 KFB38932.1 JXUM01052918 KQ561755 KXJ77622.1 GFDL01008843 JAV26202.1 SPP89967.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
UP000008237
+ More
UP000000311 UP000037069 UP000053097 UP000268350 UP000095300 UP000192221 UP000008792 UP000002358 UP000000803 UP000008711 UP000007801 UP000007798 UP000005205 UP000001070 UP000279307 UP000242457 UP000005203 UP000215335 UP000075809 UP000078492 UP000007755 UP000078540 UP000078541 UP000092553 UP000095301 UP000078542 UP000015103 UP000076502 UP000092462 UP000053825 UP000092444 UP000001819 UP000235965 UP000007266 UP000078200 UP000027135 UP000007062 UP000009046 UP000008820 UP000069272 UP000075902 UP000075885 UP000076408 UP000069940 UP000249989 UP000183832 UP000075900 UP000002320 UP000075920 UP000030765
UP000000311 UP000037069 UP000053097 UP000268350 UP000095300 UP000192221 UP000008792 UP000002358 UP000000803 UP000008711 UP000007801 UP000007798 UP000005205 UP000001070 UP000279307 UP000242457 UP000005203 UP000215335 UP000075809 UP000078492 UP000007755 UP000078540 UP000078541 UP000092553 UP000095301 UP000078542 UP000015103 UP000076502 UP000092462 UP000053825 UP000092444 UP000001819 UP000235965 UP000007266 UP000078200 UP000027135 UP000007062 UP000009046 UP000008820 UP000069272 UP000075902 UP000075885 UP000076408 UP000069940 UP000249989 UP000183832 UP000075900 UP000002320 UP000075920 UP000030765
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JBR4
A0A2H1WLR9
A0A3S2NML4
X2L6G3
A0A194QE46
A0A212F6N0
+ More
A0A194QQU0 E2B7R3 A0A1L8DMB5 E2BXN9 E2AZN9 A0A0C9RQG9 A0A0L0C373 A0A0A1WGQ3 A0A026WDG5 A0A3B0KVR9 A0A1I8PYB0 A0A1I8PYE2 W8ASL5 A0A1W4VSQ9 B4M333 K7J6I5 M9PGG4 B3P8S9 P18824 A0A034VUY2 A0A0K8U2B4 B3MXH2 B4N1V6 A0A158ND49 B4JWY3 A0A3L8E4M5 A0A2A3E2T1 A0A088ASV7 A0A232EPB5 T1PFC0 A0A151X3X9 A0A195DUF5 F4X7D1 A0A195BNB7 A0A195F7X9 A0A0M4EUK6 Q02453 A0A151I8E6 A0A0M4ESV3 T1HHR2 A0A023F5L4 A0A1Y1NCG6 A0A069DWH2 A0A154PF68 A0A0T6AU15 A0A1B0DKX3 A0A0L7R6W9 A0A146L5T1 D3TLQ8 A0A336M2A0 Q29I35 Q76CZ7 A0A2J7QDQ0 D6X202 A0A1A9VD85 A0A067QTQ8 Q7QHW5 E0V8W8 A0A1B6C185 A0A2M4A3U4 A0A2M3Z1N7 A0A2M4BDL7 A0A2M4A3V6 Q17GS9 A0A2M4CUX6 A0A2M4A3R4 A0A2M4A3L0 E0V8X0 A0A182FIY5 A0A0C9RCC0 A0A0A1X9P9 A0A0K8V7Z9 A0A1I8PYE7 W8BQR6 A0A182U3Z2 A0A182P393 A0A0K8W0W9 A0A182Y567 A0A1L8DLC2 A0A088AVH5 A0A182GGV2 A0A0P6IJT4 U5EXF9 A0A1J1J1N0 A0A067QU74 A0A182RHI9 B0WHS4 A0A182W5J9 A0A084VLT8 A0A182GBL0 A0A1Q3FFC2 P18824-2 A0A1W4VS30 A0A3B0K6X4
A0A194QQU0 E2B7R3 A0A1L8DMB5 E2BXN9 E2AZN9 A0A0C9RQG9 A0A0L0C373 A0A0A1WGQ3 A0A026WDG5 A0A3B0KVR9 A0A1I8PYB0 A0A1I8PYE2 W8ASL5 A0A1W4VSQ9 B4M333 K7J6I5 M9PGG4 B3P8S9 P18824 A0A034VUY2 A0A0K8U2B4 B3MXH2 B4N1V6 A0A158ND49 B4JWY3 A0A3L8E4M5 A0A2A3E2T1 A0A088ASV7 A0A232EPB5 T1PFC0 A0A151X3X9 A0A195DUF5 F4X7D1 A0A195BNB7 A0A195F7X9 A0A0M4EUK6 Q02453 A0A151I8E6 A0A0M4ESV3 T1HHR2 A0A023F5L4 A0A1Y1NCG6 A0A069DWH2 A0A154PF68 A0A0T6AU15 A0A1B0DKX3 A0A0L7R6W9 A0A146L5T1 D3TLQ8 A0A336M2A0 Q29I35 Q76CZ7 A0A2J7QDQ0 D6X202 A0A1A9VD85 A0A067QTQ8 Q7QHW5 E0V8W8 A0A1B6C185 A0A2M4A3U4 A0A2M3Z1N7 A0A2M4BDL7 A0A2M4A3V6 Q17GS9 A0A2M4CUX6 A0A2M4A3R4 A0A2M4A3L0 E0V8X0 A0A182FIY5 A0A0C9RCC0 A0A0A1X9P9 A0A0K8V7Z9 A0A1I8PYE7 W8BQR6 A0A182U3Z2 A0A182P393 A0A0K8W0W9 A0A182Y567 A0A1L8DLC2 A0A088AVH5 A0A182GGV2 A0A0P6IJT4 U5EXF9 A0A1J1J1N0 A0A067QU74 A0A182RHI9 B0WHS4 A0A182W5J9 A0A084VLT8 A0A182GBL0 A0A1Q3FFC2 P18824-2 A0A1W4VS30 A0A3B0K6X4
PDB
2Z6G
E-value=0,
Score=2599
Ontologies
GO
GO:0045296
GO:0007155
GO:0007616
GO:0045186
GO:0019900
GO:0030424
GO:0035147
GO:0035153
GO:0005915
GO:0030139
GO:0090254
GO:0035019
GO:0048526
GO:0060914
GO:0046530
GO:0007391
GO:1990907
GO:0007370
GO:0048477
GO:0045944
GO:0072659
GO:0046667
GO:0003713
GO:0014019
GO:0008134
GO:0000122
GO:0001709
GO:0007367
GO:0060070
GO:0060232
GO:0071896
GO:0016324
GO:0016342
GO:0035293
GO:0008587
GO:0098609
GO:0007472
GO:0005912
GO:0007399
GO:0000902
GO:0001745
GO:0005886
GO:0048754
GO:0035017
GO:0045893
GO:0014017
GO:0005829
GO:0005654
GO:0007507
GO:0005634
GO:0005737
GO:0016055
GO:0045294
GO:0019903
GO:0005913
GO:0035257
GO:0016021
GO:0008381
GO:0005515
GO:0005488
GO:0004871
GO:0008270
GO:0051287
GO:0005216
GO:0006811
GO:0006541
GO:0016812
GO:0016597
PANTHER
Topology
Subcellular location
Cytoplasm
Cell membrane
Cell junction
Adherens junction
Cell membrane
Cell junction
Adherens junction
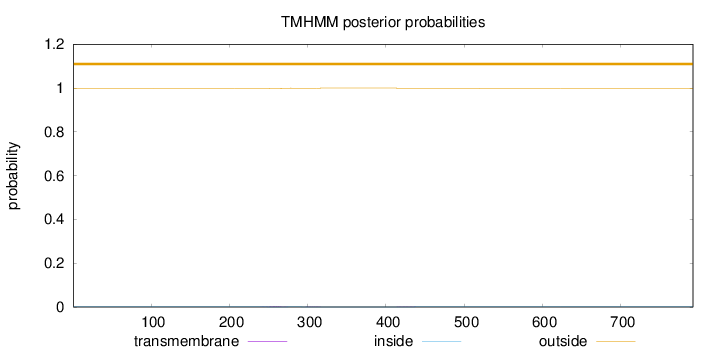
Length:
792
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0664099999999999
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00133
outside
1 - 792
Population Genetic Test Statistics
Pi
255.040805
Theta
186.891032
Tajima's D
1.034109
CLR
0.118261
CSRT
0.666516674166292
Interpretation
Uncertain
